Membrane immersion allows rhomboid proteases to achieve specificity by reading transmembrane segment dynamics
Figures
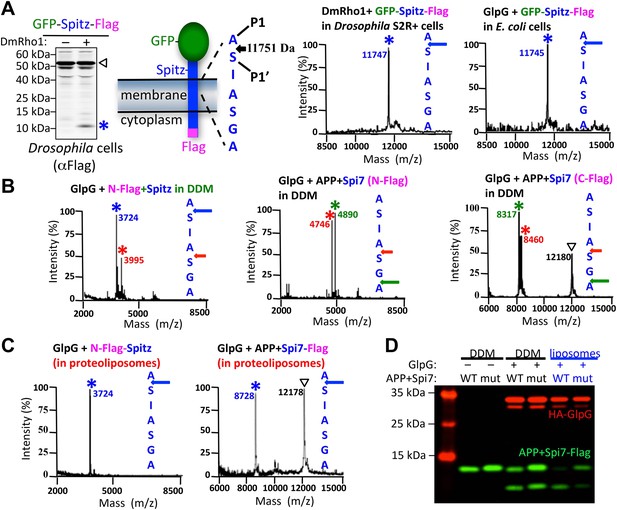
The membrane directs site and substrate specificity by rhomboid proteases.
(A) Western analysis of GFP-Spitz-Flag expressed in Drosophila S2R+ cells. Denoted throughout are uncleaved (∇) and cleaved forms (*). In vivo cleavage sites were mapped by mass spectrometry following anti-flag immunocapture of GFP-Spitz-Flag processed in Drosophila S2R+ cells by DmRho1, as well as other rhomboid proteases in both mammalian and bacterial cells and in different organelles (also see Figure 1—figure supplement 1A). The invariant cleavage site is denoted with a blue arrow in Spitz (first seven TM residues are shown). (B) The cleavage site in Spitz generated in vitro shifted also to the second AS when assayed in dodecyl-β-D-maltoside (DDM) detergent. Arrows and asterisks are color matched throughout. Cleavage products isolated from N-Flag and C-Flag tagged APP + Spi7 substrates revealed the same cleavage sites with the expected relative proportions. (C) Reconstituting substrates and rhomboid proteases from detergent into proteoliposomes in vitro restored cleavage to the natural site. (D) Cleavage of APP + Spi7-Flag vs its GA to LM mutant by GlpG in 0.25% DDM detergent or reconstituted into proteoliposomes. Note that upon reconstitution, the local concentration of substrate is higher than in detergent solution.
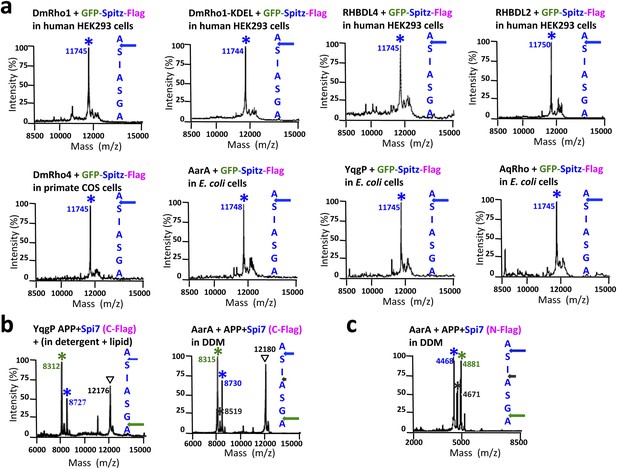
Cleavage site of Spitz in animal and bacterial cells, and APP + Spi7 in vitro.
(A) Cleavage site of GFP-Spitz-Flag processed in human HEK293 cells by DmRho1 (in the Golgi apparatus), DmRho1-KDEL (in the ER), human RHBDL4 (ER) and RHBDL2 (cell surface), DmRho4 in COS cells (cell surface), as well as in Escherichia coli cells by Providencia stuartii (AarA), Bacillus subtilis (YqgP), and Aquifex aeolicus (AqRho). Cleaved Spitz was immuno-captured with anti-Flag agarose and subjected to molecular mass analysis by MALDI-TOF mass spectrometry. The shoulder to the right of each cleaved Spitz peak results from doubly-charged immunoglobulin light chain carryover. All rhomboid proteases, irrespective of in which organelle or cell type they were expressed, cleaved Sptiz between the first two residues (AS) of its transmembrane segment. (B) Cleavage site of APP + Spi7 (C-terminal Flag tag) shifted to predominantly between the GA residues when processed by YqgP in mixed DDM:phospholipid (0.1%:0.1%) micelles (since pure YqgP is inactive in detergent alone) and AarA in detergent. Highlighted are uncleaved (∇) and cleaved forms (*). (C) Cleavage site of APP + Spi7 with AarA isolated using an N-terminal Flag tag.
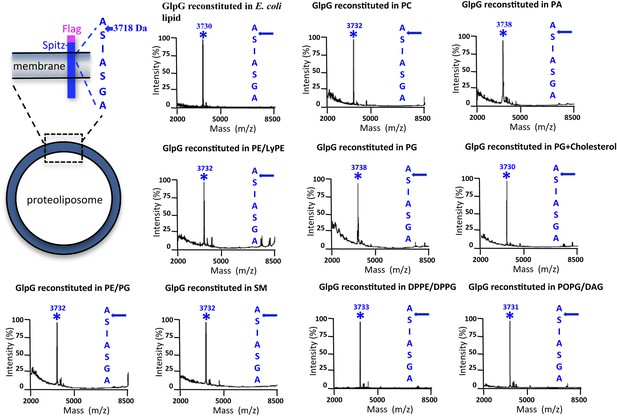
Cleavage site of N-Flag-Spitz cleaved in proteoliposomes composed of different lipids in vitro.
Sptiz was cleaved between the first two residues (AS) of its transmembrane segment irrespective of the lipid composition of the proteoliposomes. Cleaved N-terminal Flag-tagged Spitz was immuno-captured with anti-Flag agarose and subjected to molecular mass analysis by MALDI-TOF mass spectrometry. The asterisks mark the mass peak corresponding to the cleaved Flag-Spitz fragment. The lipid environments denoted are: PC, phosphatidylcholine; PA, phosphatidic acid; PE/LyPE, 1:1 molar mix of phosphatidylethanolamine and lyso-phosphatidylethanolamine; PG, phosphatidylglycerol; PE/PG, 70:30 molar mix of phosphatidylethanolamine and phosphatidylglycerol; SM, sphingomyelin; DPPE/DPPG, 70:30 molar mix of 1,2-dipalmitoyl-sn-glycero-3-phosphatidylethanolamine and 1,2-dipalmitoyl-sn-glycero-3-phosphatidylglycerol, POPG/DAG, molar 90:10 mix of phosphatidic acid and diacylglycerol; cholesterol was at 10% molar ratio.
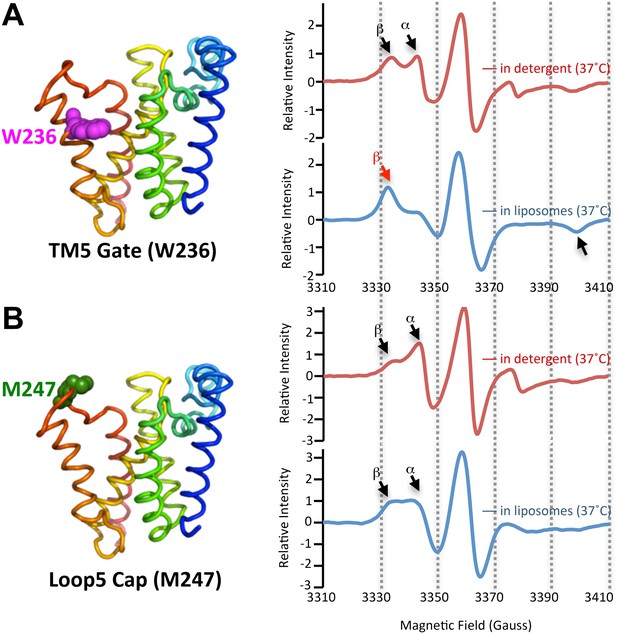
The membrane restrains rhomboid gate dynamics.
Side-view of GlpG (left) showing positions (in spheres) of nitroxide spin probes. EPR spectroscopy was conducted at 37°C in 0.5% DDM detergent or proteoliposomes formed from E. coli lipids. Shown are 100G wide first derivative spectra with the relative signal intensity between samples normalized by quantifying the absolute number of spins. Vertical dashed lines denote magnetic field value positions. (A) Dynamics at the W236 gate position in DDM detergent vs proteoliposomes: note the dramatically increased amount of the immobile β component when GlpG was analyzed in proteoliposomes (red arrow) relative to in DDM detergent. (B) Proteins dynamics at the M247 Cap position showed a major proportion in the dynamic form when GlpG was analyzed both in DDM detergent and in proteoliposomes.
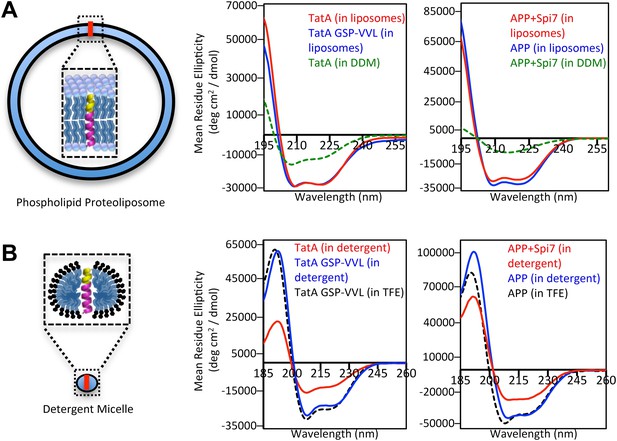
The membrane restrains substrate TM dynamics.
(A) CD spectroscopy of substrate (red traces) and non-substrate (blue traces) TM peptides revealed both display similarly stable helices when reconstituted into proteoliposomes, which was dramatically higher than in DDM detergent micelles (green dashed traces). (B) CD spectroscopy of substrate TM peptides in detergent micelles (red traces) revealed them to be strongly reduced in helicity compared to non-substrates (blue traces); in comparison, non-substrates formed helices in detergent micelles similar in stability to those in the helix-inducing solvent TFE (black dashed traces). All values are mean residue ellipticity, with relative peptide concentrations determined by simultaneously monitoring actual peptide bond absorbance during CD scanning. The TatA GSP-VVL mutant is G11V + S12V + P13L.
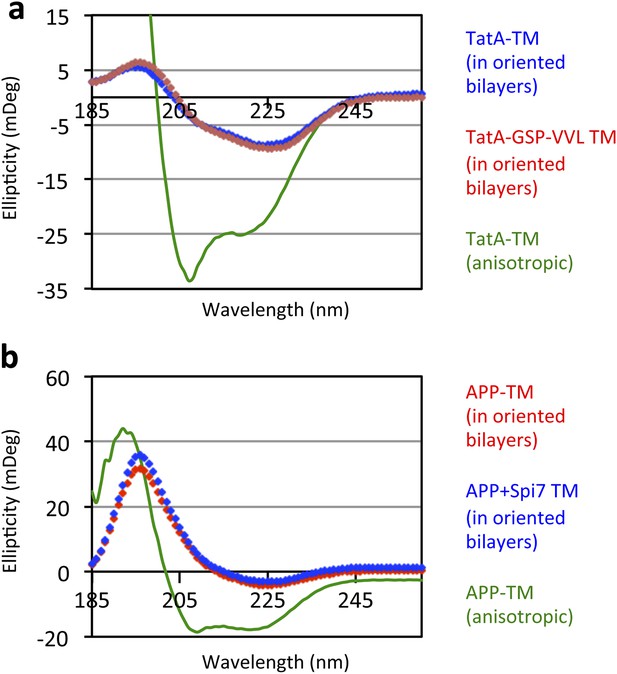
Oriented CD Spectroscopy of wildtype and mutant APP and TatA TM segments in phospholipid bilayers.
TatA (A) and APP (B) TM peptides in oriented bilayer sheets were analyzed by CD spectroscopy. Oriented CD spectra are shown in diamonds, with the red designating non-substrate TMs and blue designating rhomboid substrate TMs. The observed oriented spectra are consistent with TM peptides adopting a transbilayer orientation with no obvious differences in tilt angle between wildtype and mutant pairs. Although anisotropic, the green spectra (TM peptides in trifluoroethanol) are provided to illustrate the expected pattern for TM peptides tilted a full 90° relative to the membrane normal. Oriented bilayers at a 80:1 lipid to peptide molar ratio were assembled as described in Qian S, Wang W, Yang L, Huang HW. 2008. Biophysical J 94:3512–22.
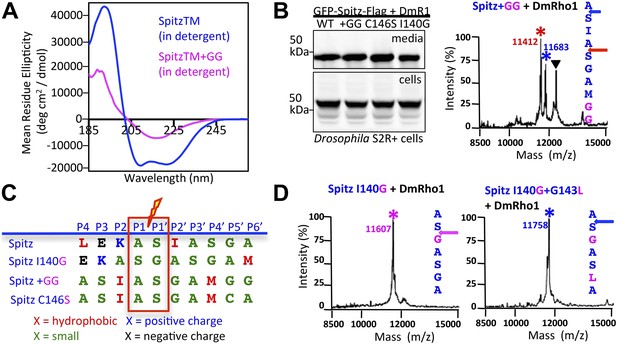
Substrate dynamics position the cleavage site by Drosophila rhomboid-1 in living cells.
(A) CD spectroscopy revealed that incorporating two glycines (+GG) near the middle of the Spitz TM further reduces its helical stability. (B) Western analysis of Spitz mutant processing by DmRho1 in living Drosophila cells assessed by amount of Spitz released into the media (top left panel). In vivo cleavage sites of Spitz + GG revealed a +3 site shift in cleavage. Black triangles denote IgG light chain from the immunoisolation. (Figure 4—figure supplement 1 shows cleavage of Spitz C146S, and a +5 site shift in cleavage of Spitz + GG by GlpG in E. coli cells). (C) Alignment of different Spitz mutant cleavage sites generated by DmRho1 in living cells. All mutants were cleaved efficiently (in B), yet note the dramatic redistribution of residues at P4, P3, P2, and P2′ positions as cleavages sites shifted. (D) The cleavage site of the Spitz I140G mutant shifted +1 residues with Drosophila rhomboid-1. Mutating the distal, helix-destabilizing G143 residue to leucine shifted cleavage site −1 residues outwards.
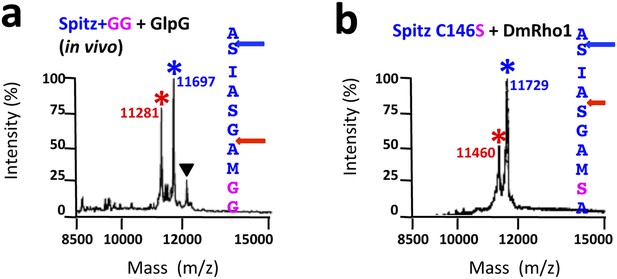
TM protein dynamics position the cleavage site.
(A) Incorporating two glycine residues (+GG) deep within the Spitz TM segment resulted in a +5 shift in cleavage site with GlpG in E. coli cells. (B) Mutating a single cysteine at position 146 to serine deep within the Spitz TM segment and far away from the cleavage site (at P8′) resulted in a +5 shift in cleavage site with Drosophila rhomboid-1 in living cells.
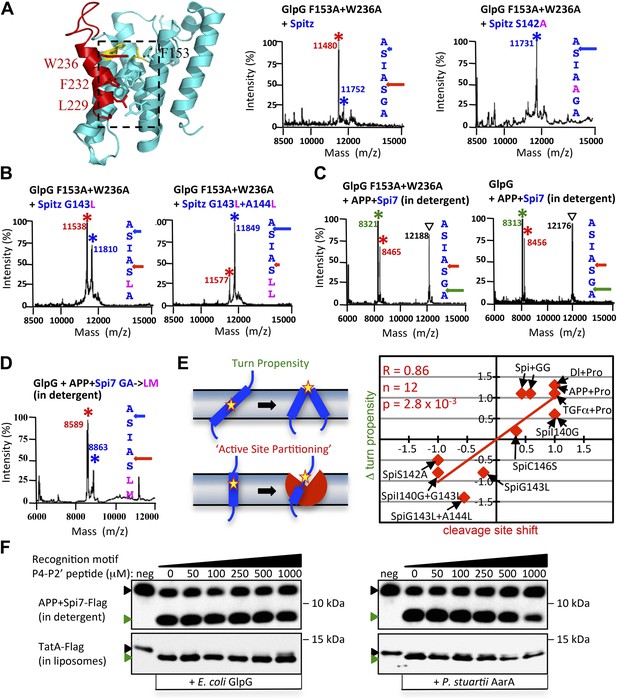
Rhomboid gate and substrate dynamics position the cleavage site by bacterial rhomboid proteases.
(A) Lateral view of GlpG showing TM5 interfacing sidechains (boxed) whose mutation opens the substrate gate (red) and increases protease activity. Gate-open mutants of GlpG analyzed in E. coli shifted cleavage site deeper into the Spitz TM segment (red). Cleavage of Spitz with helix-destabilizing S142 mutated to alanine by gate-open GlpG in E. coli cells produced a complete shift towards the top of the TM segment (blue). (B) Cleavage of Spitz with G143, and G143 + A144, mutated to leucine by gate-open GlpG in E. coli cells produced a gradual shift in cleavage site outwards. (C) Wildtype and gate-open (F153A + W236A) GlpG proteases produced identical, deeper cleavage sites when assayed in detergent micelles, indicating that the gate is fully open in the absence of the membrane. (D) Mutating the distal GA residues to LM also shifted the cleavage of APP + Spi7 in DDM detergent micelles to the natural AS site. Under these conditions the gate of wildtype GlpG is ‘open’ by virtue of the membrane being absent. (E) Diagram illustrating the ‘turn propensity’ effect of incorporating a helix-destabilizing/membrane-exiting residue (asterisk) into a long TM segment (Monné et al., 1999b). Lower diagram proposes an analogous effect for intramembrane proteolysis: residues of high ‘turn propensity’ could promote lateral substrate partitioning into the rhomboid active site. Right: change in turn propensity of substrate mutants plotted against the change in cleavage site occurring in natural membranes of living cells. (F) A hexapeptide encompassing the entire recognition motif (P4–P2′) of P. stuartii TatA, the most efficient rhomboid substrate, failed to block cleavage of two different substrates by any rhomboid tested in detergent or reconstituted liposomes (see Figure 5—figure supplement 2 for APP + Spi7 cleavage in liposomes and TatA cleavage in detergent). Black and green triangles denote substrate and cleavage product bands, respectively. The highest tested peptide concentration was 1 mM, while substrates were maintained at ≤1 μM.
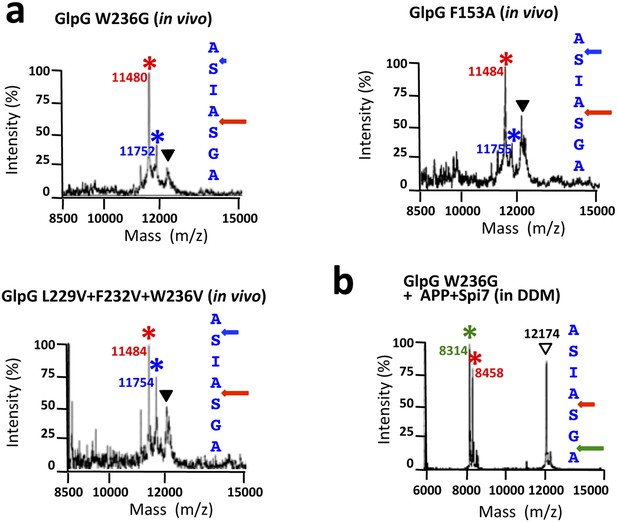
Cleavage site shifts with gate-open mutants.
(A) Cleavage site of Spitz by the gate-open TM5 mutant W236G, TM2 mutant F153A, and triple TM5 mutant L229V + F232V + W236V, in living E. coli cells shifted deeper into the transmembrane segment between the second AS pair for all gate-open mutants irrespective of their position. (B) The gate-open W236G mutant of GlpG assayed in 0.1% DDM detergent produced identical, deeper cleavage sites as the wildtype enzyme (see Figure 5C).

Poor competitive inhibition of proteolysis by a 1000-fold excess of a recognition motif peptide.
A hexapeptide (Ac-IATAAF-amide) corresponding to the entire recognition motif (P4–P2′) of TatA, the most efficient substrate for any rhomboid protease, did not block substrate proteolysis in vitro even at extreme (1 mM) concentrations. Black and green triangles denote substrate and cleavage product bands, respectively. The highest tested peptide concentration was 1 mM, while substrates were maintained at ≤1 μM. Also see Figure 5F.
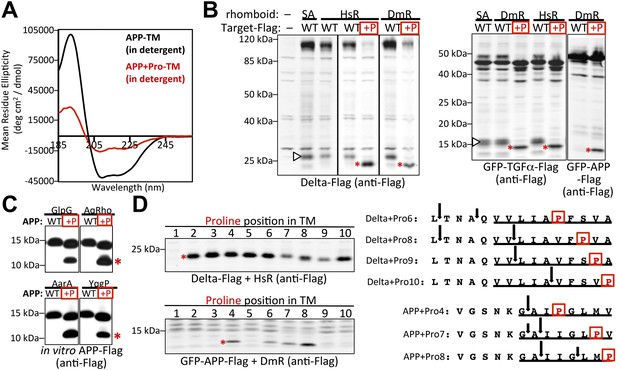
A single proline converts non-substrates into rhomboid substrates.
(A) CD spectroscopy revealed that incorporating a single proline into the TM of APP (at position 7) dramatically reduces its helical stability. (B) Incorporating a single proline into the TM segment of Delta, TGFα, or APP converted each into an efficient substrate for rhomboid proteases (DmR is Drosophila rhomboid-4, HsR is human RHBDL2, SA is the catalytic serine mutant of DmR). (−) shows non-specific anti-Flag bands in untransfected HEK293 cells. The natural juxtamembrane cleavage product generated by metalloprotease shedding (denoted by a white triangles) was outcompeted by intramembrane cleavage. (C) Incorporating a single proline also converted APP-Flag into an efficient substrate in vitro with pure bacterial rhomboid enzymes (cleaved products denoted with asterisks). (D) Effect of proline position on rhomboid proteolysis (shown are cleaved product bands, highlighted with asterisks). Cleavage sites of Delta + Pro-Flag and GFP-APP + Pro-Flag with different proline positions were mapped from living HEK293 cells (right panel). TM segment residues are underlined and the exogenous proline is boxed.
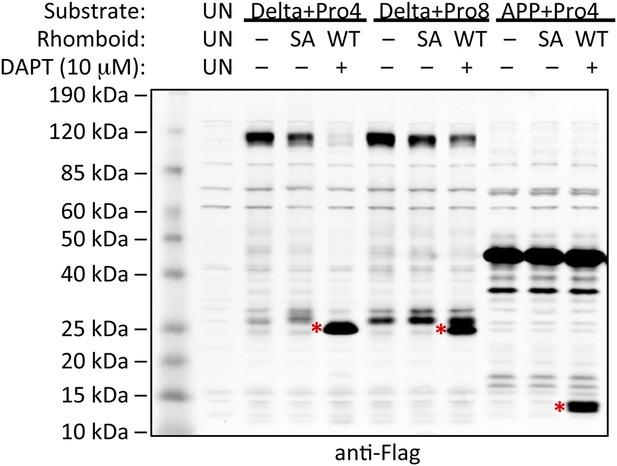
Induction of non-substrate cleavage by rhomboid proteases.
Cleavage of non-substrates harboring transmembrane proline depends on rhomboid protease activity not γ-secretase. We examined both the human (above shown with Delta) and Drosophila (above shown with APP) rhomboid proteases. ‘SA’ denotes co-transfection of HEK293 cells with rhomboid protease constructs harboring the catalytic serine mutated to alanine. These catalytically dead constructs failed to induce cleavage (highlighted with red asterisks). DAPT was added to a final concentration of 10 μM, which is approximately 100-fold higher than its reported IC50 against γ-secretase, yet it did not block proteolysis. ‘UN’ designates control untransfected cells, which serve to identify cross-reactive background bands.
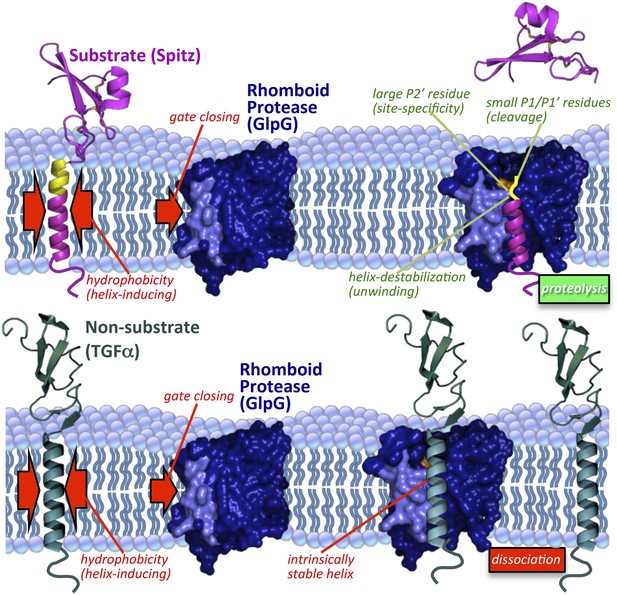
Model of rhomboid proteolysis driven by intramembrane protein dynamics.
The membrane imposes two constraints on protein dynamics to ensure high proteolytic specificity; it induces helix formation of TM segments (left red arrows) and limits rhomboid gate (light blue) opening (right red arrow). Substrates form a stable helix only in the membrane; partial exposure to the aqueous environment within rhomboid triggers an entropy-driven conformational switch, facilitated by helix-destabilizing residues, allowing substrates to reach the catalytic residues (in orange). Bottom panel depicts a non-substrate:rhomboid complex, in which the TM segment maintains a stable helix and therefore cannot reach the catalytic residues. The non-substrate TM segment eventually dissociates without being cleaved (far right). Induction of efficient non-substrate cleavage suggests that the initial docking interaction between rhomboid and TMs is non-specific. The exact order of events, and what triggers each step, remain speculative. Membrane thinning surrounding GlpG as observed in molecular dynamics simulations is illustrated (Bondar et al., 2009; Zhou et al., 2012), although its functional consequence remains unclear. Structures 2IC8 (closed GlpG), 2NRF (open GlpG), 1MOX (Spitz-EGF), and 2TGF (TGFα-EGF) were used to diagram the model.





