Engineered proteins detect spontaneous DNA breakage in human and bacterial cells
Figures
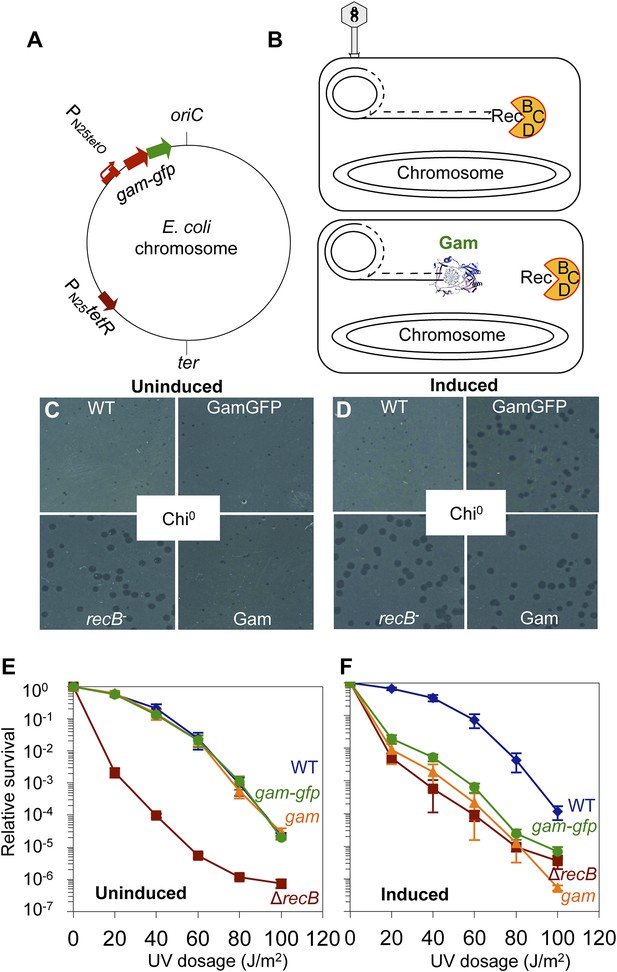
GamGFP production mimics recB double-strand-exonuclease defect.
(A) Doxycycline-inducible gam-gfp fusion construct in the E. coli chromosome. Constitutively produced TetR protein represses the PN25tetO promoter, which produces GamGFP upon doxycycline induction. oriC, origin of replication; ter, replication terminus; arrows, directions of transcription. (B) Phage λ assay for end-blocking activity by Mu Gam and GamGFP. Rolling-circle replication of phage λred gam is inhibited by E. coli RecBCD, which causes small plaques of λred gam on wild-type E. coli (Smith, 1983). Mu Gam protein binds and protects DNA ends from RecBCD exonuclease activity (Akroyd and Symonds, 1986) and so is expected to allow rolling-circle replication of λred gam and therefore allow formation of large plaques. (C) λred gam plaques are small on recB+ (WT) and large on recB-deficient cells (recB-). Plaques produced on WT cells carrying gam and gam-gfp are small when Gam and GamGFP proteins are not produced (Uninduced). (D) λred gam produce large plaques on WT cells if Gam or GamGFP are produced (Induced). (E) UV sensitivity of E. coli recB-null mutant compared with recB+(WT), and uninduced gam and gam-gfp carrying cells. WT ( ), recB− (
), recB− ( ), WT GamGFP, (
), WT GamGFP, ( ); WT Gam, (
); WT Gam, ( ). (F) Induction of Gam or GamGFP with 200 ng/ml doxycycline causes UV sensitivity similar to that of recB-null mutant cells, indicating that Gam or GamGFP block RecBCD action on double-stranded DNA ends. WT, SMR14327; recB, SMR8350; WT GamGFP, SMR14334; WT Gam, SMR14333. Representative experiment performed three times with comparable results.
). (F) Induction of Gam or GamGFP with 200 ng/ml doxycycline causes UV sensitivity similar to that of recB-null mutant cells, indicating that Gam or GamGFP block RecBCD action on double-stranded DNA ends. WT, SMR14327; recB, SMR8350; WT GamGFP, SMR14334; WT Gam, SMR14333. Representative experiment performed three times with comparable results.
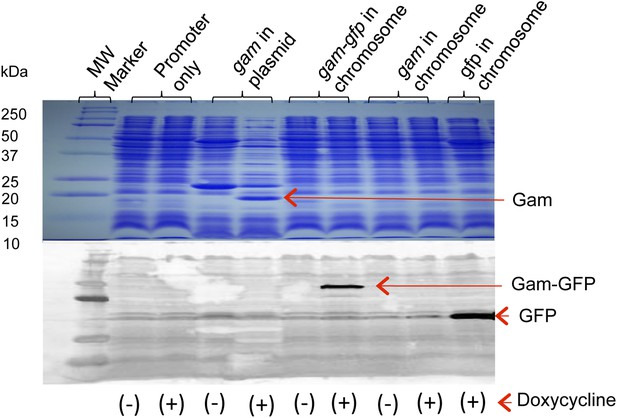
Production of Gam and GamGFP fusion proteins in E. coli.
Doxycycline induction and detection of plasmid-borne Gam and chromosomally encoded GamGFP and GFP are performed by Coomassie blue staining following electrophoresis (Gam) or by western blot immunodetection (GamGFP, GFP), in upper and lower panels, respectively. Cultures grown, as described in ‘Materials and methods’, were incubated in the presence or absence (+ or −) of 100 ng/ml doxycycline. For the western blot, protein was visualized using antibodies against GFP. Arrows indicate positions of Gam, GFP, and GamGFP within the gel. Molecular weights of protein standards are indicated to left. Strains are: promoter only, SMR14311; gam in plasmid, SMR13908; chromosomal gam-gfp, SMR14334; chromosomal gam, SMR14333; and chromosomal gfp, SMR14332.
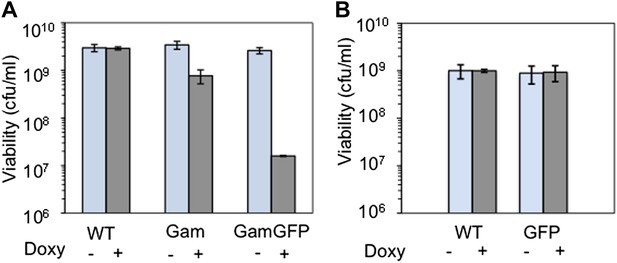
Long-term GamGFP production reduces E. coli viability.
Greater viability loss with GamGFP than Gam implies that GamGFP is a superior DSE trap. (A) We quantified the effect of long-term Gam and GamGFP production on cell viability by inducing Gam or GamGFP production briefly in split log-phase cultures, then plating them for viable colony-forming units (cfu) on inducing or non-inducing solid medium with long-term overnight incubation. Saturated LBH cultures of SMR14327 (WT), SMR14333 (Gam) and SMR14334 (GamGFP) were diluted 1:100 in fresh LBH medium and grown shaking at 37°C for 90 min, then either induced with 200 ng/ml of doxycycline or not, and grown for an additional 2 hours shaking at 37°C prior to plating for cfu on LBH solid medium with or without 200 ng/ml doxycycline. The colonies were scored after overnight incubation at 37°C. We observe that induced cultures of the Gam- and GamGFP-producing strains show, respectively, 32 ± 9% and 0.47 ± 0.06% the number of viable cfu as either WT cells with no gam gene or uninduced gam- or gam-gfp-containing cells (mean ± SD, three experiments). These data imply that DSBs bound by GamGFP are not repaired or are repaired inefficiently. Whereas the viability of Gam producers is similar to that of liquid cultures of E. coli recBC DSB-repair-defective cells, which typically contain ∼30% viable cells (e.g., Miranda and Kuzminov, 2003), GamGFP producers have lower viability. These data suggest that GamGFP is a more permanent DSE-trap and blocker of repair than Gam is, and that there is residual RecBC-independent DSB repair in recBC-defective cells. We speculate that the GFP moiety might confer more permanence to the GamGFP binding of DSBs either because the GamGFP protein inherently possesses a reduced dissociation constant or perhaps because the GFP moiety instigates multimerization with other GFP moieties in other GamGFP molecules present in the cell. This could both confer its outstanding focus-forming ability and might additionally retard end dissociation. (B) We find no cfu-reducing effect of production of GFP alone.
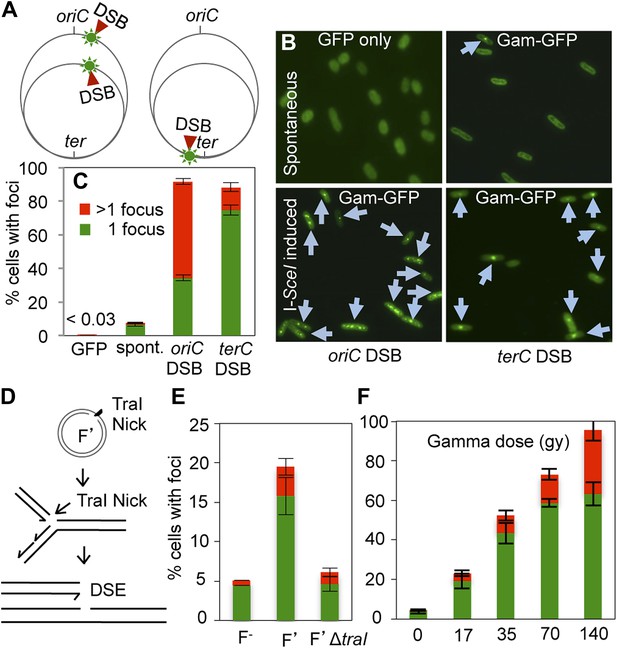
GamGFP foci at DSBs in living E. coli.
(A) Strategy. In log-phase replicating E. coli, cells have more copies of origin (oriC)-proximal than terminus (ter)-proximal DNA and so will have more DSBs per cell when cleaved by chromosomally encoded I-SceI (Ponder et al., 2005) at a cutsite (red arrow/green flash) near ori than near ter. (B) Representative data (arrows indicate foci). (C) Quantification from multiple experiments shows correlation of GamGFP foci with numbers of DSBs per cell. Cells have >1 focus when cleaved by I-SceI near ori, usually 1 focus per cell when cleaved by I-SceI near ter, far fewer cells with foci when only spontaneous DSBs are present (no I-SceI cleavage), and <0.03% of cells with foci when GFP alone is produced. E. coli strains: GFP only, SMR14332; GamGFP, SMR14350; oriC DSB, SMR14354; ter DSB, SMR14362. Error bars, ± SEM. (D) Strategy: a site-specific ssDNA nick made by TraI ssDNA endonuclease at oriT in the F plasmid becomes a one-ended DSB upon replication by fork collapse (Kuzminov, 2001). (E) TraI-dependent GamGFP foci imply that GamGFP detects one-ended DSBs. Cells with no F plasmid (F−), an F′ plasmid encoding TraI (F′), or an isogenic traI-deleted F′ (F′ΔtraI): strains SMR14015, SMR16387, and SMR16475. (F) GamGFP foci are correlated with dose of DSB-producing γ-radiation. Figure 2—figure supplement 2A shows linear correlation of foci with dose. Strain, SMR14350. Cells with 1 focus, green; >1 focus, red.
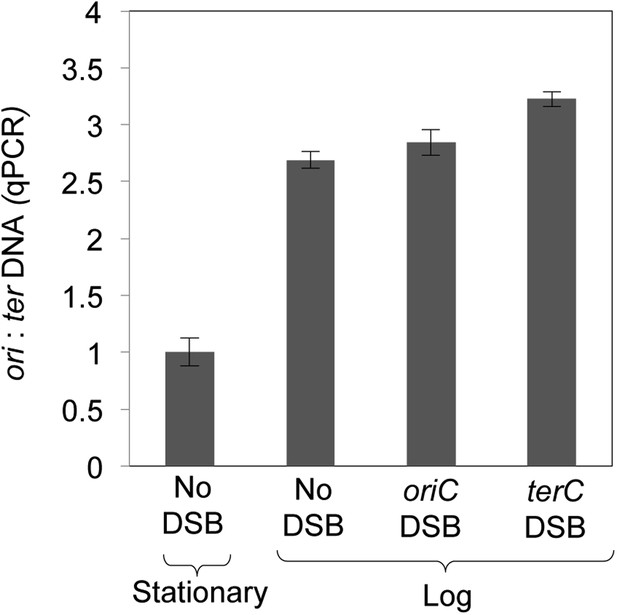
Quantitative real-time PCR shows ∼three-fold more DNA copies near ori than ter in log-phase, regardless of I-SceI cleavage, implying that some cells have two and some have four ori:ter regions.
https://doi.org/10.7554/eLife.01222.007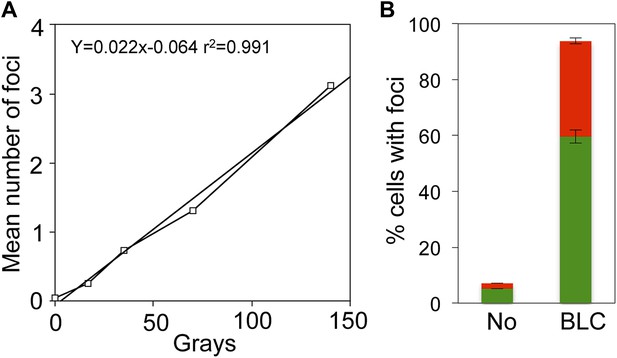
Linear gamma-ray dose-response and bleomycin induction of GamGFP foci in E. coli.
(A) Numbers of GamGFP foci are linearly correlated with dose of DSB-producing γ-irradiation. Numbers of foci at different doses of γ-irradiation were calculated from the data displayed in Figure 2F. (B) GamGFP foci form in response to bleomycin induced DSBs in living E. coli. Twenty µg/ml bleomycin (BLC) promotes GamGFP foci. Green, 1 focus per cell; red, >1 focus per cell.
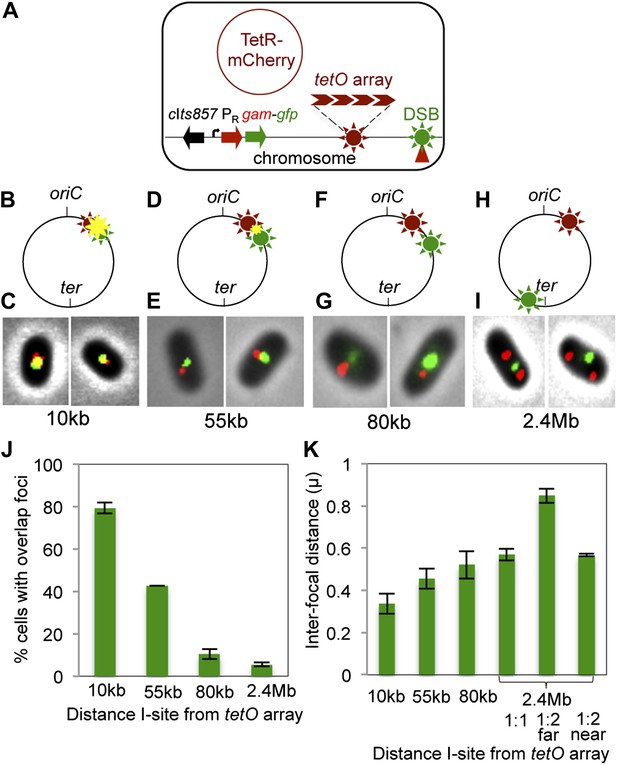
Subcellular/subgenomic localization of DSBs in living E. coli.
(A) Strategy: we varied the location of I-SceI cleavage sites (I-sites) in different strains relative to a fixed-position chromosomal TetR-mCherry-bound tetO array, with GamGFP temperature inducibly produced from chromosomal λPR (cIts857 PRgam-gfp). Red circle, plasmid that produces TetRmCherry. (B–H) Diagrams of E. coli chromosomes with inducible I-SceI endonuclease and I-sites engineered (B) 10 kb, (D) 55 kb, (F) 80 kb, and (H) 2.4 Mb from the tetO array. Co-localization of TetR-mCherry ( ) and GamGFP foci (
) and GamGFP foci ( ) results in a yellow focus (
) results in a yellow focus ( ). (C, E, G, I) Representative fluorescence microscopy results show co-localization of mCherry and GamGFP (yellow foci) at 10 kb (C), and non-overlapping foci at 55 kb (E), 80 kb (G), 2.4 Mb (I) in strains SMR16600, SMR16711, SMR16713, and SMR16606. (J) Percentage of cells with yellow overlapped foci at each distance. (K) Mean interfocal distances. At 2.4 Mb, there were frequently two red foci per one green focus, reflecting more copies of ori- than ter-proximal DNA during replication. The greater interfocal distance (far) is plotted separately from the shorter (near), and cells with 1:1 ratios were counted separately. Data represent three independent experiments, error bars indicate SEM, with the number of cells counted in all three totaling: 298, 10 kb; 204, 55 kb; 333, 80 kb; and 1347, 2.4 Mb.
). (C, E, G, I) Representative fluorescence microscopy results show co-localization of mCherry and GamGFP (yellow foci) at 10 kb (C), and non-overlapping foci at 55 kb (E), 80 kb (G), 2.4 Mb (I) in strains SMR16600, SMR16711, SMR16713, and SMR16606. (J) Percentage of cells with yellow overlapped foci at each distance. (K) Mean interfocal distances. At 2.4 Mb, there were frequently two red foci per one green focus, reflecting more copies of ori- than ter-proximal DNA during replication. The greater interfocal distance (far) is plotted separately from the shorter (near), and cells with 1:1 ratios were counted separately. Data represent three independent experiments, error bars indicate SEM, with the number of cells counted in all three totaling: 298, 10 kb; 204, 55 kb; 333, 80 kb; and 1347, 2.4 Mb.
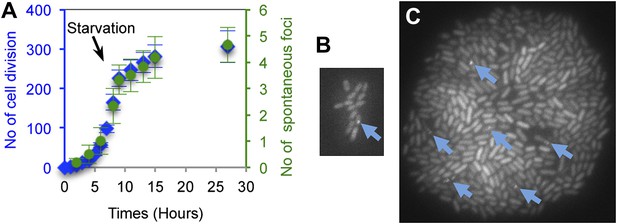
Generation-dependence of spontaneous GamGFP focus formation in proliferating E. coli.
Log-phase GamGFP-pre-induced cells were loaded into a microfluidic chamber in which single cells anchor then divide to form single-cell-layer microcolonies. The numbers of cell divisions and appearance of spontaneous foci were captured with time-lapse photography. Rapid growth in glucose during the first 9 hr was followed by washing cells in the same medium lacking glucose for 18 hr to slow and halt cell divisions. (A) Spontaneous DSB foci are correlated with numbers of cell divisions. Summary of data for six cells that became microcolonies. Blue ( ), number of cell divisions; green (
), number of cell divisions; green ( ), cumulative number of spontaneous foci that appear in each microfluidics micro-colony (mean ± SEM, six microcolonies). (B) Representative 2-hr micro-colony with a GamGFP focus (arrow). (C) Representative 15-hr micro-colony with GamGFP foci (arrows).
), cumulative number of spontaneous foci that appear in each microfluidics micro-colony (mean ± SEM, six microcolonies). (B) Representative 2-hr micro-colony with a GamGFP focus (arrow). (C) Representative 15-hr micro-colony with GamGFP foci (arrows).
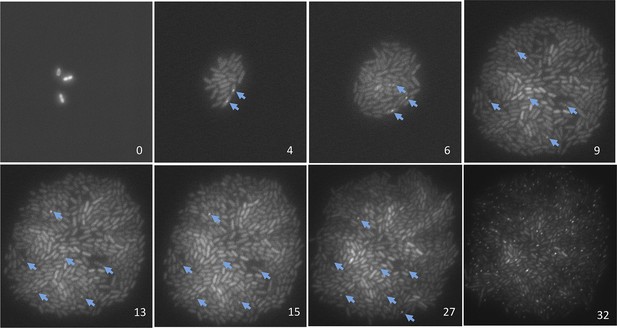
Representative data on the origins of spontaneous DSBs over time during growth, or growth retardation, visualized and quantified per ‘Materials and methods’, Microfluidics and time-lapse fluorescence microscopy of E. coli.
Numbers in the lower right of each frame are hours after loading into the microfluidic chamber. GamGFP foci are indicated with arrows. Note that cells expressing gam-gfp show variation in the amount of GFP per cell documented for E. coli (Elowitz et al., 2002) and other cells expressing any fluorescent-protein gene. This variation represents stochastic variation in transcription and mRNA accumulation, but the data scored are foci (arrows). Cells were bathed in medium with glucose for 9 hr to allow log-phase growth, then cell divisions slowed and ultimately halted (Figure 4A) by switching to the same medium without glucose. Then at 27 hr, 20 µg/ml phleomycin was added to induce DSBs, visible as foci in 45± 5% of cells at 32 hr, to verify that had DSBs formed in the starving cells, they would have been visible as foci. These images were taken under very low-dose (30 ms) exposure to fluorescent light to minimize fluorescence-induced DNA damage (Ge et al., 2013) and possible induction of GamGFP foci. Control experiments summarized in ‘Materials and methods’ show that these brief pulses did not induce GamGFP foci (Microfluidics and time-lapse microscopy of E. coli, ‘Evidence that fluorescence exposure did not contribute to the spontaneous GamGFP foci scored)’.
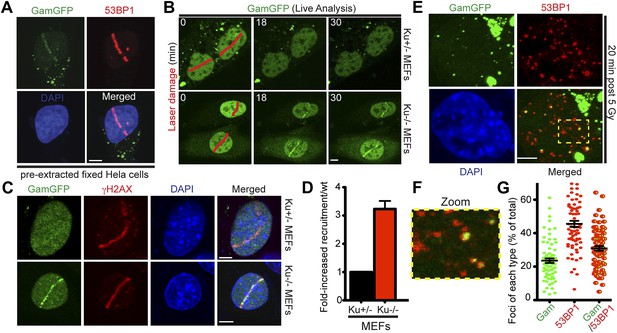
GamGFP marks DSBs in mammalian cells and is inhibited by Ku.
(A) GamGFP co-localizes with 53BP1 on laser-induced DNA breaks. (B) Ku inhibits recruitment of GamGFP to laser-induced damage, live cells. (C and D) Ku inhibits recruitment of GamGFP, fixed cells. Mean ± SEM of three experiments, n >25 cells each. (E) GamGFP forms IR-induced foci in Ku80-defective MEFs. (F) Zoomed image from E. (G) IR-induced foci containing Gam only, 53BP1 only or both Gam and 53BP1 (>2600 total foci counted in three independent experiments). Error bars, SEM. Scale bars = 5 μm.
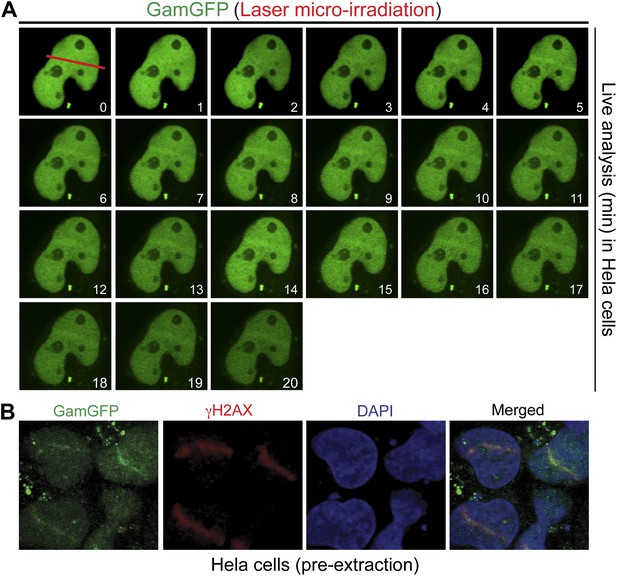
GamGFP marks DSBs in mammalian cells.
(A) Live analysis of GamGFP localization to laser-induced DNA damage. Hela cells producing GamGFP were laser damaged along the cell track indicated by the red line at 0 min (m) and images were taken at the indicated times as shown. (B) GamGFP co-localizes with γH2AX in fixed Hela cells.
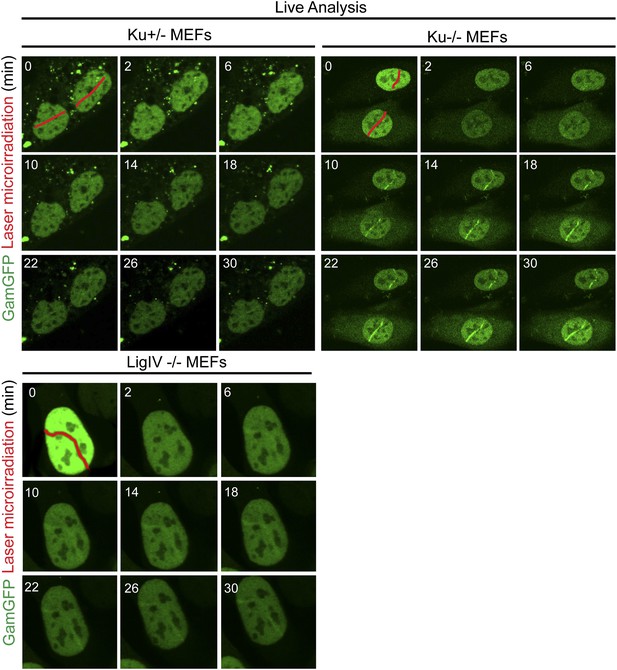
Ku inhibits GamGFP recruitment at DSBs independently of non-homologous end joining.
Cells lacking either Ku80 or LigIV are defective in non-homologous end joining (NHEJ), yet the presence of Ku still inhibits recruitment of GamGFP to laser-induced DSBs even in NHEJ-defective cells, and thus independently of the cell's ability to complete NHEJ. Whereas it could have been possible that reduced GamGFP recruitment in the presence of Ku was caused by reduced persistence of DSBs due to their repair by NHEJ, our data show instead that Ku inhibits recruitment independently of successful NHEJ and imply that Ku binding to DSEs itself is inhibitory.
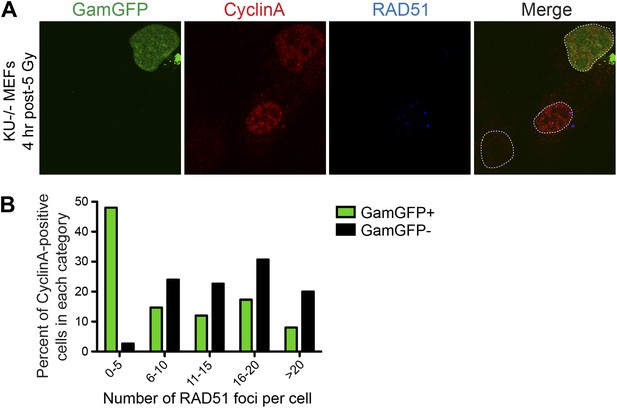
GamGFP inhibits IR-induced RAD51 foci, apparently blocking end resection.
We quantified RAD51 foci (single-stranded DNA) (Raderschall et al., 1999) induced by IR, so presumably at DSBs, in S-G2 (CyclinA-positive) cells that either did or did not produce GamGFP, from the same transfections. (A) The GamGFP-positive Ku80-defective MEFs display reduced RAD51 foci upon IR treatment in S/G2 cells. Cells were analyzed by immunofluorescence with the indicated antibodies. S/G2 cells were identified by positive staining of CyclinA. Dotted white lines mark cell nuclei. (B) Quantification of RAD51 foci in CyclinA-positive cells with or without GamGFP production (cumulative values from three experiments with >75 cells total). Each cell is ‘Z-stacked’ (optically sectioned) so that all RAD51 foci were examined. The data indicate that most GamGFP-positive cells have few (0–5) RAD51 foci per cell, and that those cells with more RAD51 foci (classes 6–10, 11–15, 16–20 and >20) are enriched among the GamGFP-negative cells. These data indicate a partial mutual exclusivity of GamGFP presence and RAD51 foci, as would be expected if Gam binding to DSEs blocks the resection that creates the ssDNA onto which RAD51 binds.
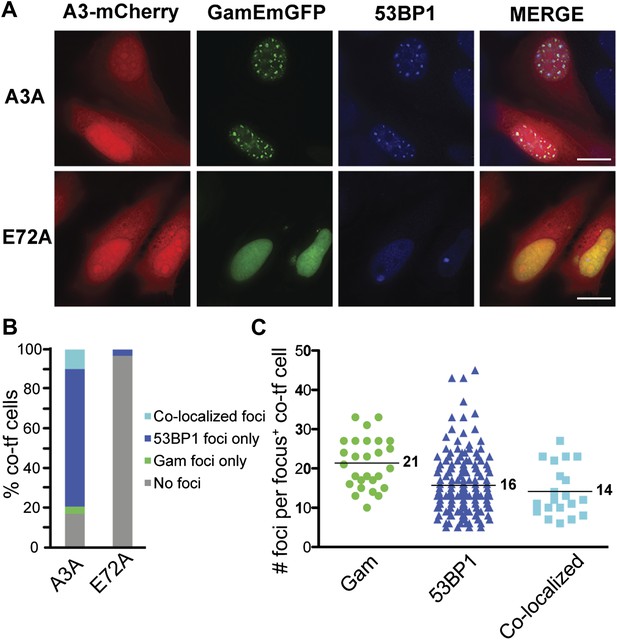
APOBEC3A induces DSBs in human cells.
(A) HeLa cells co-transfected with GamEmGFP and APOBEC3A-mCherry or catalytic mutant, APOBEC3A-E72A-mCherry. (B) Summary of foci observed in cells producing both GamEmGFP and A3A-mCherry or A3A-E72A-mCherry (two independent experiments; n = 100 per experiment). Data are percent of co-transfected cells. (C) Mean number of foci per focus-positive cell co-transfected with and expressing both GamEmGFP and A3A-mCherry.
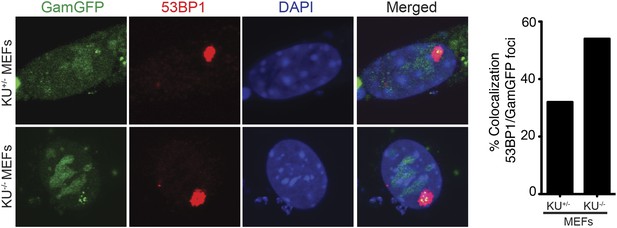
Spontaneous DNA breakage in G1-phase cells: GamGFP shows large spontaneous G1 53BP1 foci to contain multi-break clusters.
The large spontaneous 53BP1 foci in undamaged cells, which occur solely in G1 (Harrigan et al., 2011; Lukas et al., 2011a), contain multiple DSBs that are marked by GamGFP. The GamGFP-53BP1 co-localization is more apparent in the absence of Ku. Data are from three (Ku80-proficient) or four (Ku80-defective) independent experiments of >25 cells each with Z-stacked (optically sectioned) nuclei.
Additional files
-
Supplementary file 1
(A) Escherichia coli K12 plasmids and strains used in this study. E. coli strains and plasmids were constructed using standard P1 transduction (Miller, 1992), transformation, lysogenization (Gumbiner-Russo et al., 2001), recombinant DNA (Sambrook and Russell, 2001) and phage λ Red-mediated short-homology recombineering methods (Datsenko and Wanner, 2000). Sequencing was performed by SeqWright DNA Technology Services (Houston, TX). (B) PCR primers.
- https://doi.org/10.7554/eLife.01222.018





