Bacterial Growth: The SagA of E. faecium
Bacteria and other microbes residing in our intestinal tract – known as gut microbiota – are important for maintaining health and normal immunity. Among these is a group of lactic acid-producing bacteria called Enterococcus that inhibit the growth of harmful pathogens and aid digestion. As a result, these species are routinely found in probiotic supplements and fermented foods that attempt to improve gut health.
Enterococci can also become opportunistic pathogens capable of causing infections. Indeed, two of the most frequently identified species of Enterococci – E. faecium and E. faecalis – are both known to acquire antibiotic resistance. However, some strains of E. faecium have also been found to positively impact human health. For example, enrichment of E. faecium has been associated with an improved response to various types of cancer immunotherapy (Matson et al., 2018; Routy et al., 2018). Presence of these bacteria has also been shown to help prevent infections in the gut, and E. faecium are widely used as safe probiotics (Bhardwaj et al., 2010; Ishibashi et al., 2012).
E. faecium protects the gut from infections by releasing a hydrolase enzyme called secreted antigen A (SagA), which is involved in remodeling its cell wall. SagA breaks down peptidoglycans, the main component of the E. faecium cell wall, to produce small fragments called muramyldipeptides (MDPs; Kim et al., 2019). These MDPs activate receptors known as NOD2 in the host’s immune cells, leading to improved immunity in the gut.
Recent studies in mouse models showed that the MDPs generated by SagA could also increase anti-tumor immunity and improve cancer immunotherapy outcomes (Griffin et al., 2021). However, the role SagA plays in the bacteria itself remained unknown as it was believed that E. faecium needed this protein to survive. Now, in eLife, Howard Hang and colleagues – including Steven Klupt, Kyong Tkhe Fam and Xing Zhang as joint first authors – report that SagA does not affect the viability of E. faecium, but is required for cell wall remodeling and cell separation during replication (Klupt et al., 2024).
The team (who are based at Scripps Research) genetically modified E. faecium to generate a strain in which the gene for SagA was deleted. The growth of this bacterial strain was then compared to: (i) a wild-type strain, (ii) a mutant strain with inactive SagA, and (iii) a ‘complementation’ strain in which the gene for SagA had first been deleted and then re-expressed. Bacteria that lacked the gene for SagA or had an inactive version of the enzyme grew more slowly in liquid culture than wild-type E. faecium; but this was restored in the complementation strain. Experiments also showed that deleting the gene for SagA made E. faecium more sensitive to various antibiotics that target the bacterial cell wall. These exciting results raise the possibility of targeting SagA and other peptidoglycan hydrolase enzymes to overcome antibiotic resistance.
Transmission electron microscopy (TEM) – which uses a beam of electrons to generate images with ultrahigh resolution – revealed that E. faecium strains lacking the gene for SagA had more difficulty separating during replication. This caused the bacteria to cluster together, which could impair their growth. Cryo-electron tomography – a modification of TEM which can create three dimensional images of cells – was then used to quantify cell morphology parameters such as thickness of the cell wall and septum (a transient structure which helps to separate dividing cells). This revealed that deleting the gene for SagA alters the placement and projection angle of the new cell wall; however, this morphology was restored in the complementation strain.
To investigate the functional implications of deleting the gene for SagA, Klupt et al. used mass spectrometry to analyze specific components of the cell wall. They found that the strain in which the gene for SagA has been deleted generated fewer MDPs than wild-type E. faecium, which led to poor NOD2 signaling. Mouse models of cancer also did not respond to immunotherapy when they were colonized with the deficient strain. Finally, Klupt et al. demonstrated that deleting the gene for SagA reduced the population of cancer-targeting immune cells within the tumor.
Taken together, the findings show that catalytically active SagA is required for cell wall remodeling and cell separation in E. faecium, and its production of MDPs is required to mount an effective anti-tumor immune response (Figure 1). Furthermore, deleting the gene that codes for SagA impairs bacterial growth and increases sensitivity to antibiotics. Similar observations have been made in other bacteria that express peptidoglycan hydrolase enzymes, albeit via different mechanisms (Frirdich and Gaynor, 2013). This suggests that these enzymes could potentially be important antibacterial targets.
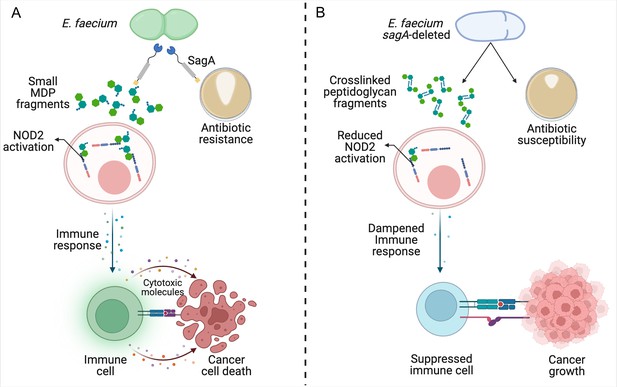
Reduced remodeling of bacterial cell walls impacts the immune response to cancer.
(A) SagA is an enzyme that helps to remodel the cell wall of E. faecium by breaking down its main component, peptidoglycan. This makes the bacteria more likely to become resistant to antibiotics (brown circle; right), allowing them to grow and form larger colonies (white shape within brown circle). SagA breaks the peptidoglycan layer into small fragments called muramyldipeptides (MDP; green hexagons), which activate NOD2 receptors in the host’s immune cells (pink). This improves the outcomes of cancer immunotherapy by triggering other cells in the immune system (green) to recognize cancer cells through receptors on the cell surface (blue and purple rectangles) and release inflammatory cytotoxic molecules that will kill them. (B) Deleting the gene for SagA (blue cell) impairs peptidoglycan remodeling and cell separation during cell division and increases the susceptibility of E. faecium to antibiotics that target the cell wall, resulting in less bacterial growth and smaller colonies. The reduced cell wall remodeling results in peptidoglycan fragments remaining crosslinked, making them too large to potently activate NOD2. This lack of sufficient NOD2 signaling prevents the immune system from mounting an appropriate immune response, leading to poorer outcomes from cancer immunotherapy.
© 2024, BioRender Inc. Figure 1 was created with BioRender.com, and published under a CC-BY-NC-ND license with permission. Further reproductions must adhere to the terms of this license.
Notably, hydrolases from other immunotherapy-promoting Enterococcus species share more than 90% sequence homology in their catalytic domain. Future studies investigating how bacteria regulate the activity of these hydrolases could lead to better treatments for cancer and combating antibiotic resistance.
References
-
Safety assessment and evaluation of probiotic potential of bacteriocinogenic Enterococcus faecium KH 24 strain under in vitro and in vivo conditionsInternational Journal of Food Microbiology 141:156–164.https://doi.org/10.1016/j.ijfoodmicro.2010.05.001
-
Peptidoglycan hydrolases, bacterial shape, and pathogenesisCurrent Opinion in Microbiology 16:767–778.https://doi.org/10.1016/j.mib.2013.09.005
-
Purification and characterization of multiple bacteriocins and an inducing peptide produced by Enterococcus faecium NKR-5-3 from Thai fermented fishBioscience, Biotechnology, and Biochemistry 76:947–953.https://doi.org/10.1271/bbb.110972
Article and author information
Author details
Publication history
- Version of Record published: April 5, 2024 (version 1)
Copyright
© 2024, Prasad and Jenq
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 472
- views
-
- 55
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Immunology and Inflammation
Environmental air irritants including nanosized carbon black (nCB) can drive systemic inflammation, promoting chronic obstructive pulmonary disease (COPD) and emphysema development. The let-7 microRNA (Mirlet7 miRNA) family is associated with IL-17-driven T cell inflammation, a canonical signature of lung inflammation. Recent evidence suggests the Mirlet7 family is downregulated in patients with COPD, however, whether this repression conveys a functional consequence on emphysema pathology has not been elucidated. Here, we show that overall expression of the Mirlet7 clusters, Mirlet7b/Mirlet7c2 and Mirlet7a1/Mirlet7f1/Mirlet7d, are reduced in the lungs and T cells of smokers with emphysema as well as in mice with cigarette smoke (CS)- or nCB-elicited emphysema. We demonstrate that loss of the Mirlet7b/Mirlet7c2 cluster in T cells predisposed mice to exaggerated CS- or nCB-elicited emphysema. Furthermore, ablation of the Mirlet7b/Mirlet7c2 cluster enhanced CD8+IL17a+ T cells (Tc17) formation in emphysema development in mice. Additionally, transgenic mice overexpressing Mirlet7g in T cells are resistant to Tc17 and CD4+IL17a+ T cells (Th17) development when exposed to nCB. Mechanistically, our findings reveal the master regulator of Tc17/Th17 differentiation, RAR-related orphan receptor gamma t (RORγt), as a direct target of Mirlet7 in T cells. Overall, our findings shed light on the Mirlet7/RORγt axis with Mirlet7 acting as a molecular brake in the generation of Tc17 cells and suggest a novel therapeutic approach for tempering the augmented IL-17-mediated response in emphysema.
-
- Immunology and Inflammation
SARS-CoV-2 vaccines have been used worldwide to combat COVID-19 pandemic. To elucidate the factors that determine the longevity of spike (S)-specific antibodies, we traced the characteristics of S-specific T cell clonotypes together with their epitopes and anti-S antibody titers before and after BNT162b2 vaccination over time. T cell receptor (TCR) αβ sequences and mRNA expression of the S-responded T cells were investigated using single-cell TCR- and RNA-sequencing. Highly expanded 199 TCR clonotypes upon stimulation with S peptide pools were reconstituted into a reporter T cell line for the determination of epitopes and restricting HLAs. Among them, we could determine 78 S epitopes, most of which were conserved in variants of concern (VOCs). After the 2nd vaccination, T cell clonotypes highly responsive to recall S stimulation were polarized to follicular helper T (Tfh)-like cells in donors exhibiting sustained anti-S antibody titers (designated as ‘sustainers’), but not in ‘decliners’. Even before vaccination, S-reactive CD4+ T cell clonotypes did exist, most of which cross-reacted with environmental or symbiotic microbes. However, these clonotypes contracted after vaccination. Conversely, S-reactive clonotypes dominated after vaccination were undetectable in pre-vaccinated T cell pool, suggesting that highly responding S-reactive T cells were established by vaccination from rare clonotypes. These results suggest that de novo acquisition of memory Tfh-like cells upon vaccination may contribute to the longevity of anti-S antibody titers.






