TBP/TFIID-dependent activation of MyoD target genes in skeletal muscle cells
Figures
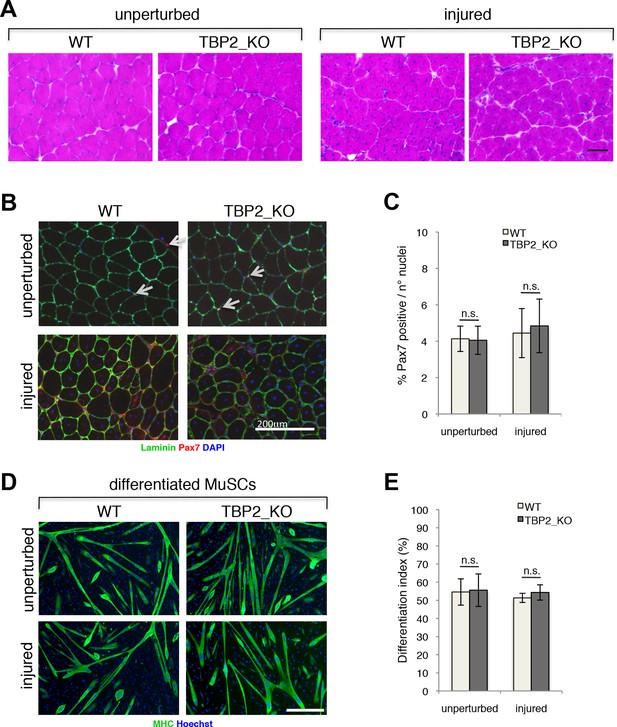
Regenerative potential and differentiation of MuSCs are intact in the absence of TBP2.
(A) Cross-sections of unperturbed (left panel) and notexin injured (right panel) muscles were histologicaly assessed by Hematoxilin/Eosin (H&E) staining to evaluate the muscle architecture 12 days post notexin-mediated injury of the muscle. Size bar 100 µm. Representative image of muscles analyzed from two WT and two TBP2_KO mice are presented. (B) Representative images of cross-sections of unperturbed and of notexin injured muscles were immunolabeled with anti-laminin B2 (green) and anti-Pax7 (red) antibodies. Nuclei were counterstained with DAPI (blue). Pax7-positive MuSCs (Muscle Stem cells - Satellite cells) in sub-laminar position are indicated by an arrow. Size bar 200 µm. (C) Quantification of Pax7-positive MuSCs residing in sub-laminar position in B. Muscles from two mice have been analyzed and for each point, three different areas of the muscle were taken into account. Error bars represent a standard deviation among biological replicates. Student’s t test was used for statistical analyses in C and E, n.s. - not significant. (D) MuSCs were isolated by FACS-assisted analysis as CD45-/CD31-/Ter119-/Sca1-/CD34+/alpha7intergrin+ from unperturbed and from notexin injured (12 days post injury) muscles. Isolated MuSCs were in vitro differentiated into myotubes, and immuno-labeled with anti-Myosin Heavy Chain (MHC) antibody (green) to monitor myotubes formation. The nuclei were counterstained with Hoechst 33,258 (blue). Images were taken by Olympus IX71 microscope with 10x objective. Representative images of differentiated MuSCs are presented. Size bar 50 µm. (E) Quantification of nuclei residing within MHC-positive differentiated MuSCs myotubes in D. Differentiation index is presented as% on nuclei within MHC-positive myotubes. Error bars represent a standard deviation between two independent MuSC isolations, each cultured and quantified in triplicate. Student’s t test was used for statistical analyses in C and E, n.s. - not significant.
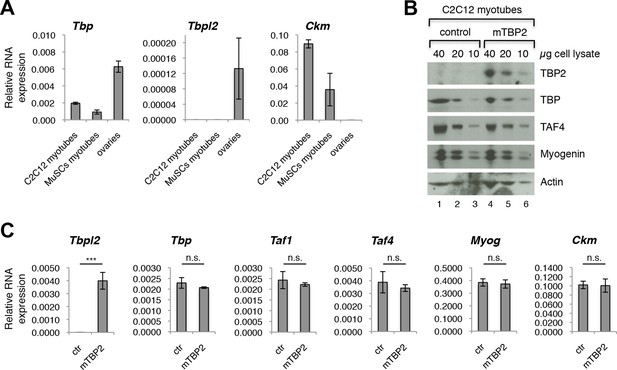
TBP2 is not expressed in myotubes.
(A) RT-PCR analysis of RNA isolated from MuSC-derived myotubes, and from C2C12-derived myotubes. RNA isolated from murine ovaries was used as a control for Tbpl2 expression (gene coding for TBP2 protein). Relative expression of indicated genes is presented as a fraction of Gapdh expression. Biological triplicates, error bars represent standard deviation. (B) Immunoblot analysis of the whole cell lysate of C2C12 myotubes exogenously expressing murine Tbpl2 gene (mTBP2) or with a control plasmid. (C) RNA expression of indicated genes after exogenous expression of TBP2 in differentiated C2C12 myotubes. Biological triplicates, error bars represent standard deviation. Student’s t test was used for statistical analyses (***p < 0.001, n.s. - not significant). Myotubes in all experiments shown in this figure were collected by careful trypsinization to avoid contamination with undifferentiated reserve cells (Kitzmann et al., 1998).
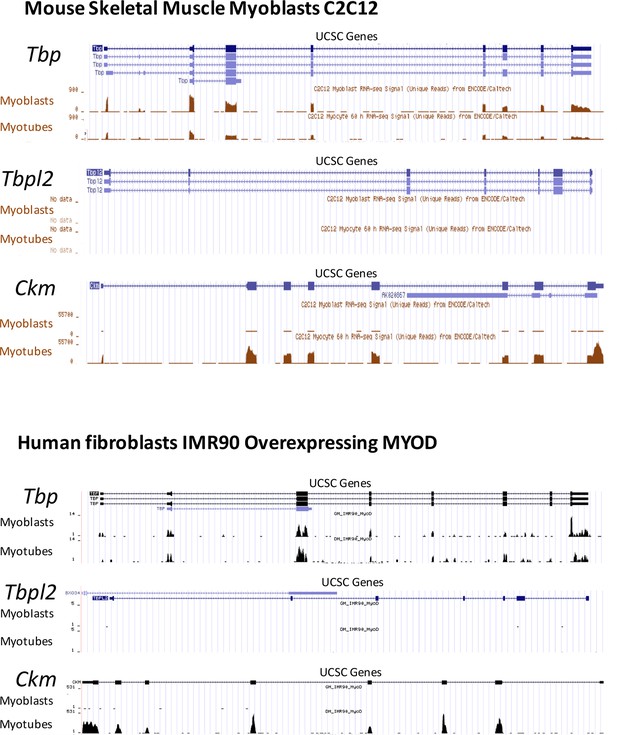
RNA-seq data comparing Tbp and Tbpl2 gene expression in murine C2C12 and in human fibroblasts IMR90 converted by ectopic expression of MyoD.
RNA-seq data in C2C12 proliferating moblasts (Myoblasts) and differentiated C2C12 (Myotubes) were generated in B. Wold's laboratory (GSE20846) (Trapnell et al., 2010). RNA-seq data in IMR90 fibroblasts expressing MyoD were generated in our lab (doi:10.5061/dryad.7qk36). GM_IMR90_MyoD sample represents proliferating IMR90 expressing MyoD (Myoblasts) and DM_IMR90_MyoD sample represents IMR90 expressing MyoD and differentiated into myotubes (Myotubes). Tbp (gene coding for TBP) and Tbpl2 (gene coding for TBP2) data are presented, Ckm is a marker of terminally differentiated myotubes. RNA-seq data were uploaded to UCSC genome browser for visual representation.
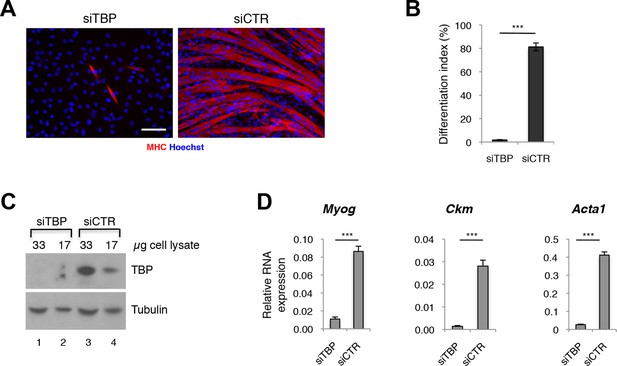
TBP is required for myoblast differentiation into myotubes.
(A) Representative images of C2C12 myoblasts differentiation into myotubes upon TBP downregulation. TBP was downregulated using Tbp-targetting siRNA in C2C12 myoblasts. C2C12 myoblasts were differentiated into myotubes for two days, and immunolabeled with anti-Myosin Heavy Chain (MHC) antibody (red) to monitor myotybes formation. The nuclei were counterstained with Hoechst 33,258 (blue). Images were taken by Olympus IX71 microscope with 20x objective. Size bar 20 µm. (B) Quantification of nuclei residing within differentiated MHC-positive myotubes in A, represented as differentiation index. Error bars represent a standard deviation among three biological replicates. Student’s t test was used for statistical analyses (***p<0.001). (C) Immunoblot analysis of the whole cell lysate of C2C12 to assess the TBP protein levels after siRNA-mediated silencing of TBP. (D) mRNA isolated from C2C12s grown in differentiation medium after siRNA-mediated downregulation of TBP was analyzed by RT-PCR for expression of skeletal muscle specific genes Myog, Ckm and Acta1. Relative expression of indicated genes is presented as a fraction of Gapdh expression. Error bars represent a standard deviation among three biological replicates. Student’s t test was used for statistical analyses (***p < 0.001).
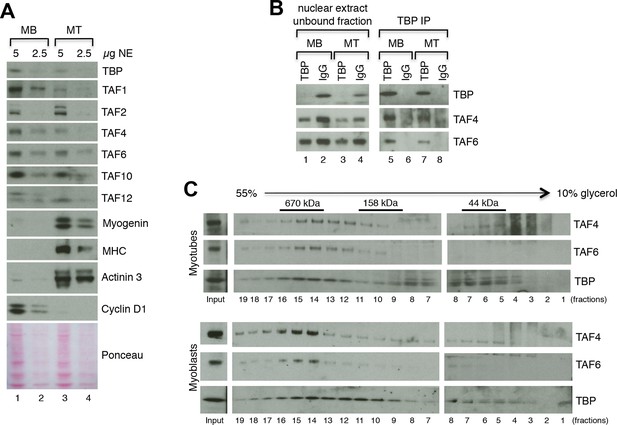
TFIID complex is present in proliferating myoblasts and differentiated myotubes.
(A) Immunoblot analysis from nuclear extracts was used to determine protein levels of TFIID subunits TBP, TAF1, TAF2, TAF4, TAF6, TAF10, TAF12 expressed in proliferating myoblasts (MB) and differentiated myotubes (MT). Myoblast-specific expression of Cyclin D1 and myotube-specific expression of muscle markers Myogenin, MHC and Actinin 3 are used as markers of C2C12 differentiation. Ponceau staining of proteins on the immunoblot membrane was used as a loading control. Representative experiment is show. (B) Immunoprecipitation of endogenous TBP from C2C12 nuclear extract results in co-immunoprecipitation of TAF4 and TAF6 subunits of TFIID in both myoblasts (MB) and myotubes (MT). (C) 10–55% glycerol gradient size fractionation assay monitors co-fractionation of TFIID subunits TBP, TAF4 and TAF6 in high molecular weight fractions with a signal peaking in fractions corresponding to 0.6–1 MDa in both myoblasts (lower panel) and myotubes (upper panel).

Evaluation of TFIID subunit expression on RNA level during C2C12 and Satellite cells (MuSCs) differentiation.
Reverse Transcription - PCR (RT-PCR) analysis of RNA isolated from proliferating and differentiated (A) C2C12 and (B) Satellite cells (MuSCs). Signal is represented as a fraction of Gapdh expression levels. Myotubes were collected by careful trypsinization (Kitzmann et al., 1998). Error bars represent a standard deviation among three biological replicates. Student’s t test was used for statistical analyses (*p<0.05, **p<0.01, ***p<0.001).
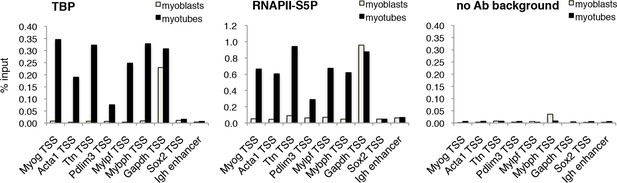
TBP is recruited to muscle genes upon muscle differentiation.
Chromatin Immunoprecipitation (ChIP) assay in proliferating C2C12 myoblasts and differentiated C2C12 myotubes monitors recruitment of TBP and RNAPII-S5P components of PIC at promoters of skeletal muscle genes Myog, Acta1, Ttn, Pdlim3, Mylpf and Mybph. Transcriptionally active Gapdh promoter and inactive Sox2 promoter and Igh enhancer were used as controls. Signal of relative recruitment is represented as a percentage of input DNA. Representative experiment of two independent biological replicate experiments is shown.
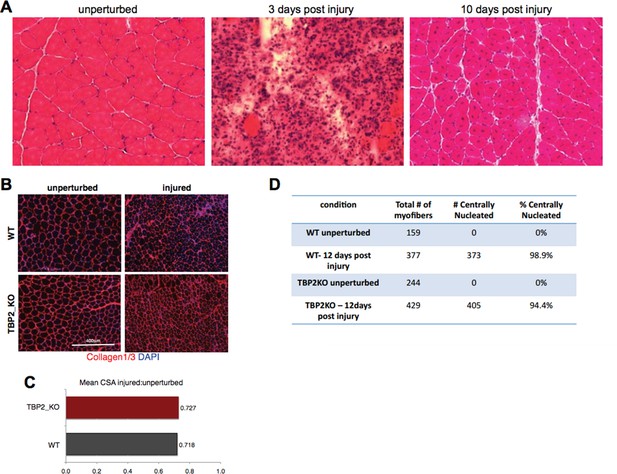
Injury decreases the CSA in regenerating myofibers to the same extend in WT and TBP2KO.
(A) Muscle histology: Hematoxilin & Eosin staining of WT muscle crosssections at different time point of regeneration. (B) Immunohistochemistry: cross-sections of WT and TBP2_KO muscles were stained with anti-collagen1 and anti-collagen-3 antibodies (red) to visualize clearly the myofibers. The nuclei were counterstained with DAPI. (C) Mean of all cross-section areas (CSA) quantified by ImageJ for each muscle was compared between injured and unperturbed muscle, the ratio is plotted. D) The centrally nucleated myofibers in 1B were quantified.
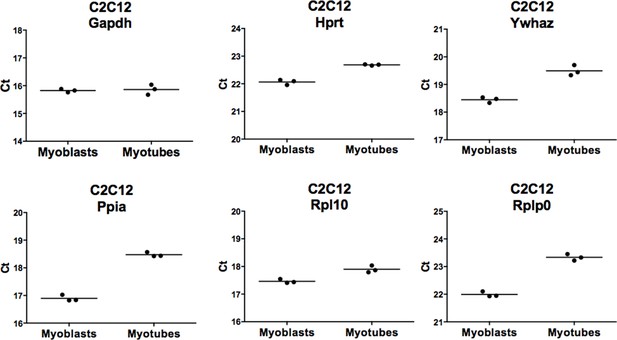
Comparison of expression levels of six houskeeping genes during myoblasts to myotubes differentiation.
RNA was isolated from biological triplicates of C2C12 myoblasts and myotubes as described in themanuscript (see Figure 4—figure supplement 1). Identical amount of RNA was converted to cDNA for each sample. RNA expression for six houskeeping genes was evaluated by RT-PCR: Gapdh,Hprt, Ywhaz, Ppia, Rpl10 and Rpl0. Ct values for each analyzed genes were plotted, the bar represents mean of three independent samples.
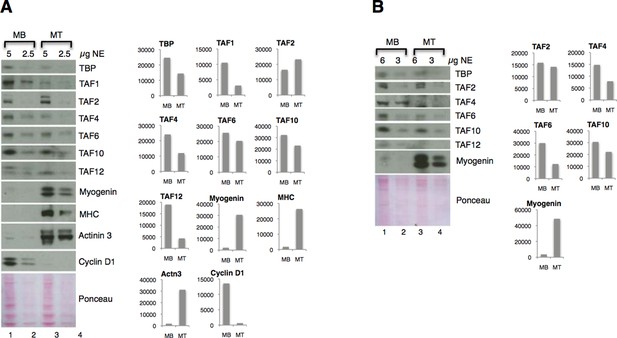
Downregulation of TFIID subunits on protein level - two independent nuclear extract (NE) preparations.
(A) Original figure in the manuscript (Figure 4A). (B) Immunoblot analysis of independent nuclear extract preparation. In panel B signals for TBP and TAF12 were not possible to quantify due to a high background:signal ratio. Immunoblot signal was quantified with ImageJ.
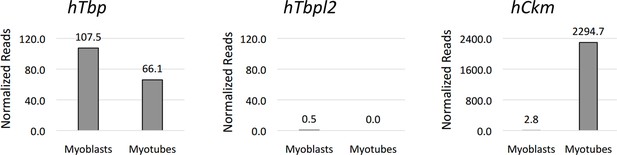
Graphical representation of normalized RNA-seq data in human IMR90 fibroblasts overexpressing MyoD grown in growth media GM (myoblasts) or differentiated in differentiation media DM (myotubes).
https://doi.org/10.7554/eLife.12534.014Tables
Squences of the primers used in this study.
| Mouse primers for RT-PCR | |
|---|---|
| Gapdh-for | GCTCACTGGCATGGCCTTCCG |
| Gapdh-rev | GTAGGCCATGAGGTCCACCAC |
| Myog-for | GAGACATCCCCCTATTTCTACCA |
| Myog-rev | GCTCAGTCCGCTCATAGCC |
| Acta1-for | AOCGTTTCCGTTGCCCOOAG |
| Acta1-rev | GGAGAGAGAGCGCGAACGCA |
| Ttn-for | GACACCACAAGGTGCAAAGTC |
| Ttn-rev | CCCACTOTTCTTGACCGTATCT |
| Pdlim3-for | TGGGGGCATAGACTTCAATCA |
| Pdlim3-rev | CTCCGTACCAAAGCCATCAATAG |
| Mylpf-for | TTCAAGGAGGCGTTCACTQTA |
| Mylpf-rev | TAOCGTCOAGTTCCTCATTCT |
| Mybph-for | CAGCCACTAAGCCTGAACCTC |
| Mybph-rev | TCCAACACATAGCCTTGAAGC |
| Cyclin D1-for | ACTTCCTCTCCAAAATGCCAG |
| Cyclin D1-rev | GTOGGTTGOAAATGAACTTCAC |
| Ckm-for | AQTCCTACACQQTCTTCAAGO |
| Ckm-rev | AGGAAGTGGTCATCAATQAGC |
| Tbpl2-for | ATACCTGGACCTCTTCCTGOAT |
| Tbpl2-rev | CCACCAAGATGTGGATGAAAC |
| Tbp-for | CCAAGCGATTTGCTGCAGTCATCA |
| Tbp-rev | ACTTAGCTGGGAAGGCCAACTTCT |
| Taf1-for | ACAGGAACAGATGCAGACCTTCGT |
| Taf1-rev | AATCTCCTCCTCAGGCACACCAAA |
| Taf2-for | GGCCTTGGAAAAATTCCCCAC |
| Taf2-rev | GAAGCACGCTGACATCCTGA |
| Taf3-for | GACATTGATGCTGCGAAAGTGCGA |
| Taf3-rev | TCCCGCTTGCT7CTTTCTCGATCT |
| Taf4-for | AGTTCACACGGCAAAGAATCACGC |
| Taf4-rev | AACGCCGGCTCATCCTGTTACTTA |
| Taf5-for | TTTC6GACGAGTAAATTCGTTCT |
| Taf5-rev | CTCCTGCACGATGTTCCAGAT |
| Taf6-for | AAACTCAGCAATACTGTGTTGCC |
| Taf6-rev | TTCTGTCGTTTCCCCATGTGC |
| Taf8-for | CCGGGAAGTAAGCAATCCACT |
| Taf8-rev | GCTTTCTCGGCACTCTCAAATC |
| Tar9-for | TGCCGAAAGATGCACAGATGA |
| Taf9-rev | TGTTGTCACATATCOGAAGGC |
| Taf10-for | GAGGGOGCAATQTCTAACGG |
| Taf10-rev | TGTGTAATCCTCCAACTGCATC |
| Taf12-for | GGACAGQTQQTCGTCTCAG |
| Tat12-rev | TCATCAOCOATCTOTAGCAGC |
| Mouse primers for ChIP | |
| Myoq TSS-for | GCTCAGGTTTCTGTGGCGTT |
| Myog TSS-rev | CCAAC TGCTGGGTGCC AT |
| Acta1 T5S-for | GTGCCCGACACCCAAATA |
| Acta1 TSS-rev | AGGGTAGGAAGTGAGGCTT |
| Ttn TSS-for | CCTTCCTAACAGAGCCAATCAC |
| Ttn TSS-rev | TGTTTCCTATGCAATCCCTACAC |
| Pdlim3 TSS-for | CACTCGCAGCAGGQATAAAT |
| Pdlim3 TSS-rev | GAACCGGACAACCTACTTAGC |
| Mylpf TSS-for | CTCCAAGCAGATTCTCTTGCTTT |
| Mylpf TSS-rev | GGTAGOGC TAT CC T OAOCTAAT |
| Mybph TSS-for | GCCTGCCTTTATAAGCATQAAC |
| Mybph TSS-rev | GTOTCAAGCTGOAGTOTTTAAG |
| Gapdh TSS-for | AGQGCTGCAGTCCGTATTTA |
| Gapdh TSS-rev | AGOAGGGGAAATGAGAOAGG |
| Sox2 TSS-for | GATTGGCCGCCGQAAAC |
| Sox2 TSS-rev | CTCTTCTCTOCCTTOACAACTC |
| Igh enhancer-for | AACCACAGCTACAAGTTTACC |
| Igh enhancer-rev | AACCAGAAÇACCTGCAGCAGC |





