The GIRK1 subunit potentiates G protein activation of cardiac GIRK1/4 hetero-tetramers
Figures
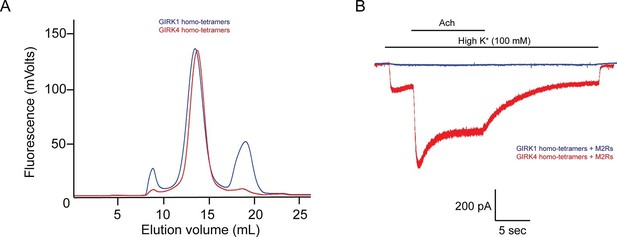
The GIRK4 subunit forms functional homo-tetrameric channels, whereas the GIRK1 subunit forms nonfunctional homo-tetramers.
(A) HEK293T cells were transiently transfected with the GIRK1 or the GIRK4 subunit fused to GFP, and solubilized cell lysate was analyzed by fluorescent size-exclusion chromatography (Superose 6 10/300 GL). Blue and red elution profiles show GIRK1 homo-tetramers and GIRK4 homo-tetramers, respectively. (B) HEK293T cells were transiently transfected with GIRK1 (blue) or GIRK4 (red), and human M2Rs. Whole-cell voltage clamp recordings were performed. Membrane potential was held at -80 mV, and the extracellular solution was exchanged to high potassium buffer (100 mM KCl) as indicated above the signal, followed by the application of 10 µM acetylcholine.
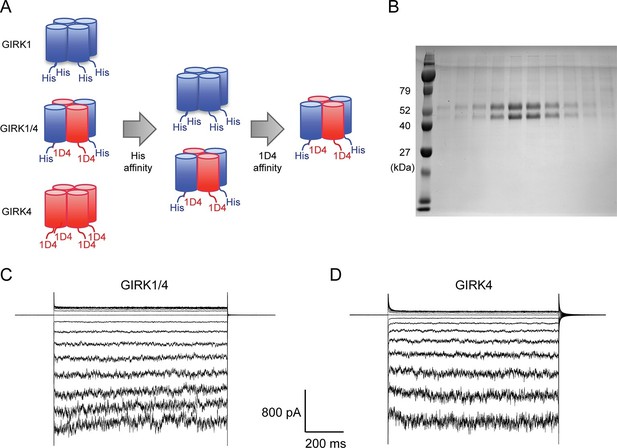
Purified cardiac GIRK channels are functional in reconstituted planar lipid bilayer membranes.
(A) Schematic of GIRK1/4 hetero-tetramer purification procedure. 1D4-tagged GIRK4 homo-tetramers were removed with Co2+ affinity chromatography and His-tagged GIRK1 homo-tetramers were removed with subsequent 1D4 affinity chromatography. (B) Gel-filtration fractions of the GIRK1/4 hetero-tetramer peak were run on 12% SDS-PAGE. GIRK1 and GIRK4 monomers are 56 kDa and 46 kDa, respectively. (C) and (D) The top and bottom chambers are separated by the lipid bilayer formed on a transparency film. The same solution containing 10 mM potassium phosphate buffer pH 7.4 and 150 mM KCl was used in both chambers. Proteoliposomes containing GIRK channels were fused to the bilayer membrane. 32 µM C8-PIP2 and 2 mM MgCl2 were added to the intracellular side of the chamber, and proteoliposomes containing Gβγ were fused to the membrane, activating GIRK channels. (C) GIRK1/4 hetero-tetramer currents recorded in the lipid bilayer. Membrane potential was held at 0 mV, and 10 mV voltage steps from -80 mV to 80 mV were applied. (D) GIRK4 homo-tetramer currents recorded in the lipid bilayer.
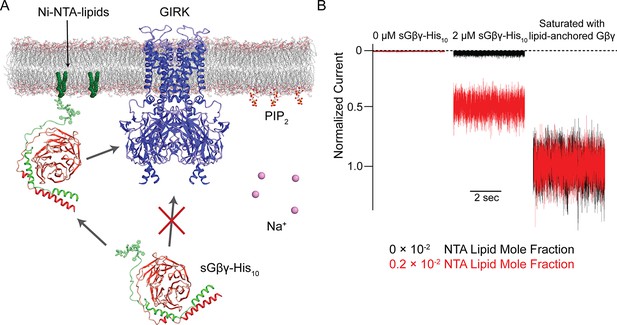
Schematic of the Na+ and Gβγ titration using Ni-NTA-lipids.
(A) GIRK channels were incorporated into the membrane containing a known concentration of Ni-NTA-lipids. 32 µM C8-PIP2 and 2 µM sGβγ-His10 were added to the intracellular side of the membrane. Free sGβγ-His10 does not activate GIRK channels at the concentration applied, while Ni-NTA-lipids-bound sGβγ-His10 mimics lipid-anchored Gβγ and activates GIRK channels. Known concentrations of Na+ were subsequently added to study the effect of Na+ concentration on GIRK channel activity in the presence of known concentrations of Gβγ in the membrane. (B) Left and center traces show normalized GIRK4 currents before and after application of 2 µM sGβγ-His10 in the presence of 0 (black) or 0.002 (red) mole fraction of Ni-NTA-lipids in the membrane. At the end of each experiment, currents were fully activated by lipid-anchored Gβγ (right signals).
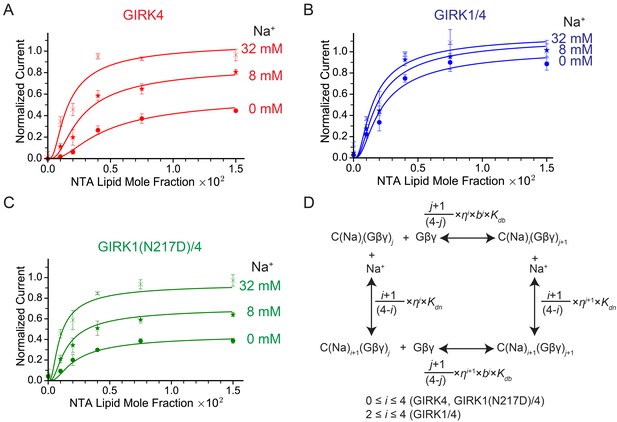
GIRK channel activity as a function of Na+ and Gβγ concentrations.
(A), (B), and (C) Plots of activity of GIRK4 homo-tetramers (A), GIRK1/4 hetero-tetramers (B), and GIRK1(N217D)/4 hetero-tetramers (C) versus Ni-NTA-lipid mole fraction in the membrane at different Na+ concentrations. The same buffer (10 mM potassium phosphate pH 8.2, 150 mM KCl) was used in both chambers, and voltage across the lipid bilayer was held at -50 mV. GIRK proteoliposomes were fused to the bilayer membrane containing a known concentration of Ni-NTA-lipids. 2 mM MgCl2 and 32 µM C8-PIP2 were added to one side of the bilayer chamber and then 2 µM sGβγ-His10 was added to the same side of the chamber to activate GIRK channels. 8 mM and 32 mM Na+ were added to further activate GIRK channels. At the end of each experiment, channels were fully activated by fusing proteoliposomes containing lipid-anchored Gβγ and currents were normalized to the fully activated current (mean ± SEM, n = 3 bilayers). The equilibrium model (D) was used to fit the data (solid curves). Kdb: Equilibrium dissociation constant between Gβγ and ligand-free GIRK. Kdn: Equilibrium dissociation constant between Na+ and ligand-free GIRK (mM). b: Cooperativity factor for Gβγ binding. η: Cross-cooperativity factor between Gβγ and Na+ binding. i: The number of Na+ ions bound to GIRK. For GIRK1/4 hetero-tetramers, the range of i was restricted to the range 2 to 4. j: The number of Gβγ bound to GIRK.
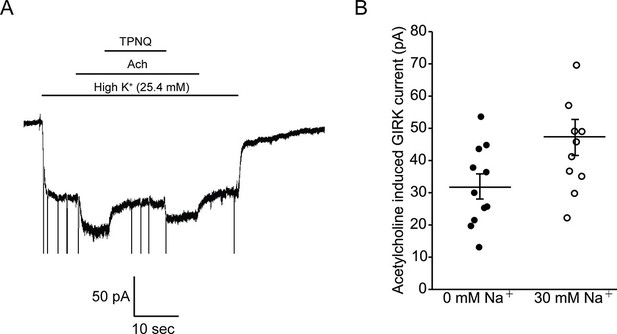
Intracellular Na+ does not significantly activate cardiac GIRK channels.
(A) Whole-cell voltage clamp recordings on mESC-derived pacemaker cells. Membrane potential was held at -80 mV and the extracellular solution was exchanged to high potassium buffer (25.4 mM KCl) as indicated above the signal. 10 µM acetylcholine (Ach) was then applied to activate GIRK channels and 100 nM tertiapin Q (TPNQ) was next applied to block cardiac GIRK currents. Acetylcholine-activated GIRK currents were measured by subtracting signals before and after acetylcholine application. (B) Acetylcholine induced GIRK currents were measured with the pipette solution containing 0 mM Na+ or 30 mM Na+. Eleven recordings were performed with each pipette and average value was calculated (mean ± SEM, n = 11 cells).
Tables
The fitting parameters for the Na+ and Gβγ titration. Kdb: Equilibrium dissociation constant for Gβγ in equilibrium with ligand-free GIRK. Kdn: Equilibrium dissociation constant for Na+ binding to ligand-free GIRK (mM). b: Cooperativity factor for Gβγ binding. η: Cross-cooperativity factor between Gβγ and Na+ binding. θi,j: Normalized activity of i-Na+ and j-Gβγ-bound GIRK. R2: Adjusted R-squared. For fitting to GIRK1(N217D)/4 hetero-tetramers, b and θ4,4 were fixed to the same parameters as GIRK1/4 hetero-tetramers.
| GIRK4 | GIRK1/4 | GIRK1(N217D)/4 | |
|---|---|---|---|
| Kdb | 0.004 ± 0.005 | 0.004 ± 0.006 | 0.0024 ± 0.0004 |
| kKdn(mM) | 50 ± 40 | 50 ± 300 | 50 ± 30 |
| b | 0.6 ± 0.3 | 0.6 ± 0.3 | 0.6 |
| η | 0.7 ± 0.1 | 0.8 ± 0.4 | 0.71 ± 0.08 |
| θ0,4 | 0.6 ± 0.1 | - | 0.45 ± 0.04 |
| θ1,4 | θ0,4 + (θ4,4- θ0,4) × 1/4 | - | θ0,4 + (θ4,4- θ0,4) × 1/4 |
| θ2,4 | θ0,4 + (θ4,4- θ0,4) × 2/4 | 1.1 ± 0.1 | θ0,4 + (θ4,4- θ0,4) × 2/4 |
| θ3,4 | θ0,4 + (θ4,4- θ0,4) × 3/4 | 1.2 ± 0.8 | θ0,4 + (θ4,4- θ0,4) × 3/4 |
| θ4,4 | 1.2 ± 0.1 | 1.1 ± 0.8 | 1.1 |
| R2 | 0.96 | 0.93 | 0.97 |





