Neural Coding: Taking a close look at electrosensing
The question of how animals make sense of their environment is of interest in both neuroscience and computer science. The key challenge is to understand how to categorize events in ways that are useful to the animal, and also practical to implement. Typically, this involves separating information about an event (for example, what kind of object has just appeared in the animal's environment?) from information about where the event happened (DiCarlo et al., 2012).
While much of the research in this area has focused on vision, similar computational principles are involved for other senses, including smell and hearing (King and Nelken, 2009). In particular it is thought that the initial signal detected by sensory neurons in the peripheral regions of the brain undergoes a series of transformations as it is passed from one set of neurons to another. The end result is to produce an representation of the event (for example, the presence of another animal of a particular species) that does not depend on where the event happened. However, much remains to be understood about the transformations that would make this possible.
In this situation, studying species with exotic senses not used by humans might shed new light on the problem. Now, in eLife, Michael Metzen, Volker Hofmann and Maurice Chacron from McGill University report the results of experiments on fish called brown ghost knifefish (Metzen et al., 2016). These fish live in murky water and produce electric signals in order to “see” their environment. Such fish are known as weakly electric fish because the electrical discharges they produce, while strong enough to "see" objects, are not strong enough to be used as weapons.
A brown ghost knifefish emits an electric field that oscillates at a given frequency and creates an electric field that surrounds the fish. It also uses an array of electroreceptors on its surface to detect changes in this electric field: these changes could be caused by other fish, both electric and non-electric, predator or prey, as well as other objects. Metzen et al. focused on what happens when a knifefish communicates with another knifefish. Each knifefish emits at a slightly different frequency, so when two knifefish come into close proximity of each other, these frequencies interfere to generate a beat pattern. The frequency of this beat pattern is equal to the difference between the two original frequencies.
If a knifefish wants to communicate with another knifefish, it produces a “chirp” – a temporary increase in the frequency of its electric field discharge (Zupanc and Maler 1993; Figure 1). This leads to a change in both the frequency and phase of the beat pattern (with the change in the phase having any value between zero and 360 degrees). The phase can change again (by between zero and 360 degrees) depending upon the timing of the response from the second fish. Given this ambiguity, how can the fish recognize each other when each fish can produce a wide range of different phase changes in the beat pattern?
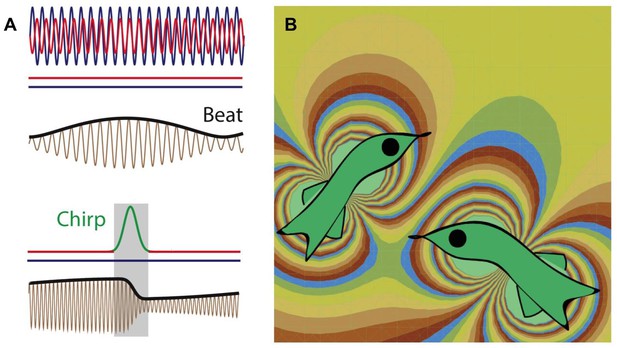
Weakly electric fish generate electric fields to communicate and to probe their environment.
(A) The electric fields produced by two electric fish (red and blue lines; top) interfere to produce a beat pattern (brown and black lines; middle); the horizontal red and blue lines indicate that the frequency of each field does not change with time. If the red fish wants to communicate with the blue fish, it increases the frequency of the electric field it produces for a short time (red-green-red line; bottom). This "chirp" changes both the frequency and phase of the beat pattern (brown and black lines; bottom). Image from Metzen et al. (B) Computer simulation (computed using the dipole approximation) showing the electric fields produced by two electric fish. The fish on the left is able to detect the presence of the fish on the right because the latter changes the electric field in the vicinity of the former, and vice versa. The different colors represent different strengths and directions of the electric field.
Metzen et al. studied how the communication signal is represented in three successive stages of neural circuits: circuits formed by neurons that are directly connected to the electroreceptors on the surface of the fish; circuits formed by pyramidal neurons in the hindbrain; and circuits formed by neurons within the Torus semicircularis in the midbrain. Metzen et al. report that neural responses at each successive stage of processing depend less and less on the phase. This process of making the signals 'phase invariant' is similar to how visual signals are processed (Movshon et al., 1978; Sharpee et al., 2013).
Studies of the comparatively simple neural circuits of the weakly electric fish can also provide insights into the neural mechanisms behind invariant responses in other more complex sensory systems. For example, the responses of the individual peripheral neurons in the knifefish are not phase invariant: however, the activity of these neurons is correlated in a way that is phase invariant. This shows that one way to generate a phase-invariant response is to correlate and combine a number of neural responses.
A prominent aspect of electrosensation is that the animal has control over the types of signals that it produces. This makes it a rich ground for testing the theory that biological circuits are optimized to transmit maximal information for a given cost (Polani, 2009; Bialek, 2013). Relevant questions include: why are the electro-receptors on the surface of the fish distributed the way they are, and why do the communication calls used by knifefish have the structure they do? And given that fish continue to grow throughout their lives, how does the electrosensory system continue to operate when the anatomy of the fish is changing constantly? An answer to this last question could help to improve our understanding of how mammalian systems cope with aging and adaptation (Webster et al., 2006).
More generally, one expects to find many solutions to the problem of information optimization, depending on the characteristics of the environmental niche for a particular animal and the metabolic costs they can afford (Tishby and Polani, 2011). The diversity of these solutions can be mapped onto biological diversity. For example, why do some fish probe their environment with electric fields, whereas bats use echolocation? From a computational perspective, there are also parallels between electrosensation in weakly electric fish and song communication in birds, auditory vocalizations in dolphins and non-human primates, and human speech (Kanwal and Rauschecker, 2007). Comparative analysis of these systems in terms of information and energy efficiency will help to elucidate the general principles of how neural circuits work.
References
-
BookInformation theory of decisions and actionsIn: Cutsuridis V, Hussain A, Taylor JG, editors. Perception-Action Cycle. New York: Springer. pp. 601–636.https://doi.org/10.1007/978-1-4419-1452-1_19
-
Auditory cortex of bats and primates: Managing species-specific calls for social communicationFrontiers in Bioscience 12:4621–4640.https://doi.org/10.2741/2413
-
Receptive field organization of complex cells in the cat's striate cortexJournal of Physiology 283:79–99.https://doi.org/10.1113/jphysiol.1978.sp012489
-
Trade-off between curvature tuning and position invariance in visual area V4Proceedings of the National Academy of Sciences of the United States of America 110:11618–11623.https://doi.org/10.1073/pnas.1217479110
-
Neural adjustments to chromatic blurSpatial Vision 19:111–132.https://doi.org/10.1163/156856806776923380
-
Evoked chirping in the weakly electric fish apteronotus leptorhynchus : A quantitative biophysical analysisCanadian Journal of Zoology 71:2301–2310.https://doi.org/10.1139/z93-323
Article and author information
Author details
Publication history
- Version of Record published: April 29, 2016 (version 1)
Copyright
© 2016, Sharpee
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 885
- Page views
-
- 115
- Downloads
-
- 0
- Citations
Article citation count generated by polling the highest count across the following sources: Crossref, PubMed Central, Scopus.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Computational and Systems Biology
Revealing protein binding sites with other molecules, such as nucleic acids, peptides, or small ligands, sheds light on disease mechanism elucidation and novel drug design. With the explosive growth of proteins in sequence databases, how to accurately and efficiently identify these binding sites from sequences becomes essential. However, current methods mostly rely on expensive multiple sequence alignments or experimental protein structures, limiting their genome-scale applications. Besides, these methods haven’t fully explored the geometry of the protein structures. Here, we propose GPSite, a multi-task network for simultaneously predicting binding residues of DNA, RNA, peptide, protein, ATP, HEM, and metal ions on proteins. GPSite was trained on informative sequence embeddings and predicted structures from protein language models, while comprehensively extracting residual and relational geometric contexts in an end-to-end manner. Experiments demonstrate that GPSite substantially surpasses state-of-the-art sequence-based and structure-based approaches on various benchmark datasets, even when the structures are not well-predicted. The low computational cost of GPSite enables rapid genome-scale binding residue annotations for over 568,000 sequences, providing opportunities to unveil unexplored associations of binding sites with molecular functions, biological processes, and genetic variants. The GPSite webserver and annotation database can be freely accessed at https://bio-web1.nscc-gz.cn/app/GPSite.
-
- Cell Biology
- Computational and Systems Biology
Computer models of the human ventricular cardiomyocyte action potential (AP) have reached a level of detail and maturity that has led to an increasing number of applications in the pharmaceutical sector. However, interfacing the models with experimental data can become a significant computational burden. To mitigate the computational burden, the present study introduces a neural network (NN) that emulates the AP for given maximum conductances of selected ion channels, pumps, and exchangers. Its applicability in pharmacological studies was tested on synthetic and experimental data. The NN emulator potentially enables massive speed-ups compared to regular simulations and the forward problem (find drugged AP for pharmacological parameters defined as scaling factors of control maximum conductances) on synthetic data could be solved with average root-mean-square errors (RMSE) of 0.47 mV in normal APs and of 14.5 mV in abnormal APs exhibiting early afterdepolarizations (72.5% of the emulated APs were alining with the abnormality, and the substantial majority of the remaining APs demonstrated pronounced proximity). This demonstrates not only very fast and mostly very accurate AP emulations but also the capability of accounting for discontinuities, a major advantage over existing emulation strategies. Furthermore, the inverse problem (find pharmacological parameters for control and drugged APs through optimization) on synthetic data could be solved with high accuracy shown by a maximum RMSE of 0.22 in the estimated pharmacological parameters. However, notable mismatches were observed between pharmacological parameters estimated from experimental data and distributions obtained from the Comprehensive in vitro Proarrhythmia Assay initiative. This reveals larger inaccuracies which can be attributed particularly to the fact that small tissue preparations were studied while the emulator was trained on single cardiomyocyte data. Overall, our study highlights the potential of NN emulators as powerful tool for an increased efficiency in future quantitative systems pharmacology studies.





