Structural basis for the inhibition of RecBCD by Gam and its synergistic antibacterial effect with quinolones
Figures
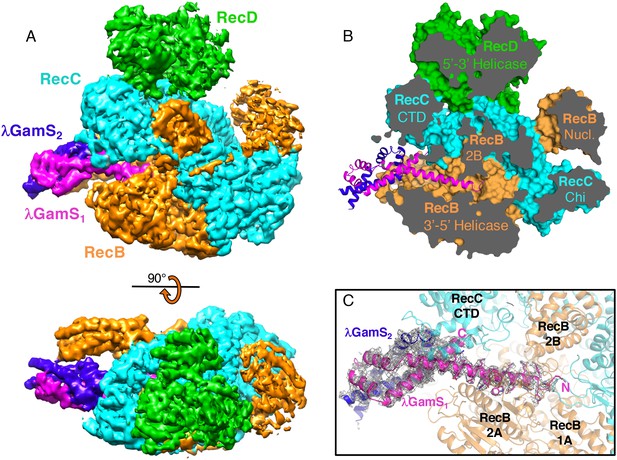
The overall structure of the RecBCD/Gam complex.
(A) Surface representation of the electron density with a ribbon representation of the RecBCD subunits and the GamS protein dimer. (B) Cut away of the molecular surface of the RecBCD part of the model with the GamS dimer overlaid showing how the protein enters and fills the channel normally occupied by the 3’ssDNA tail. (C) The same view with the electron density for the GamS dimer overlaid.
-
Figure 1—source data 1
EM data statistics and Final model.
- https://doi.org/10.7554/eLife.22963.003
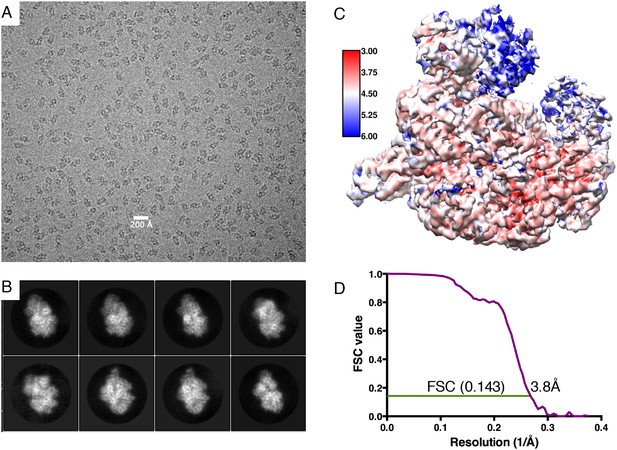
Electron microscopy information.
(A) Typical micrograph of RecBCD/GamS complex particles. (B) A selection of 2D classes during refinement. (C) Local resolution map of the final reconstruction. (D) Gold standard FSC correlation curve.
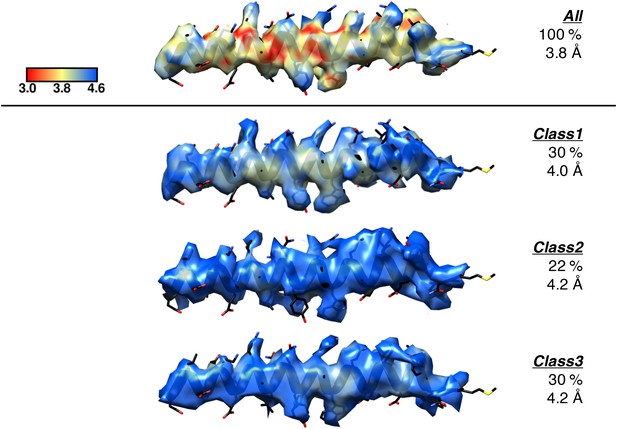
Representation of the local resolution of the GamS1 N-terminal helix for the deposited map compared to the same region in the maps of each of the three sub-classes.
For each, only the extent of the map within 2.5 Å of the region of interest was displayed over the final refined coordinates (black), with all maps contoured to the same level in Chimera (0.04). ResMap was used to colour the surface of each map according to the local resolution as outlined by the scale bar. The three sub-classes were the major classes observed when the full dataset was re-classified using 3D classification without alignment as described in Materials and methods, with the resulting percentage distributions and final refined resolution limits indicated.
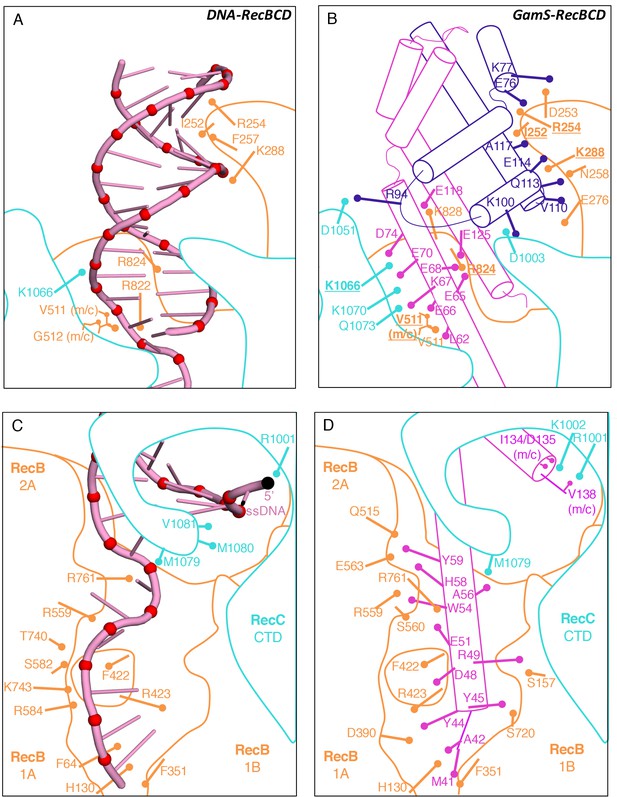
Comparison of the RecBCD/Gam and RecBCD/DNA interfaces.
(A) Cartoon representation of contacts between RecBCD and the duplex portion of bound DNA. RecB is in orange and RecC in cyan in all panels. Interactions involving main chain atoms are denoted as m/c. (B) The same interface but with the GamS dimer (shown in magenta and purple). The interaction is much more extensive than with DNA but contacts in common are shown in bold. (C) Contacts between RecBCD and the ssDNA tails of DNA. (D) The same interface but with the GamS dimer. Again the interactions are much more extensive but still include several residues in common (shown in bold).
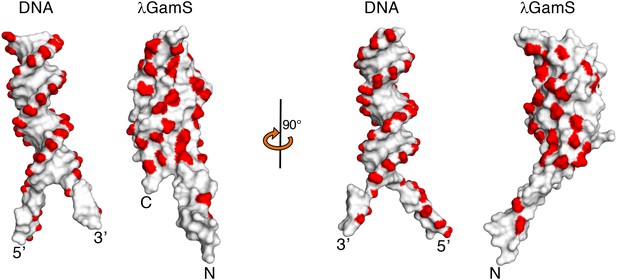
Space filling representations of the bound DNA substrate and GamS dimer with negative charges coloured in red.
https://doi.org/10.7554/eLife.22963.009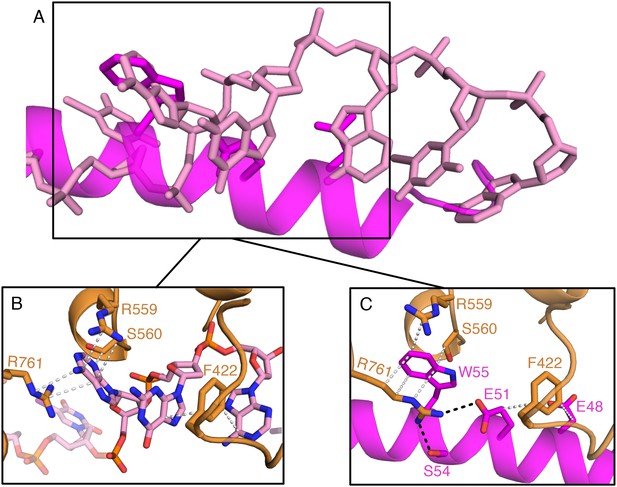
Aspects of molecular mimicry are shown in a comparison of the interactions in RecBCD/DNA complex with that of the RecBCD/Gam complex in the region of the 3’-tail of the DNA substrate.
(A) Overlap of the ssDNA (pink) and N-terminal helix of Gam (magenta). (B) The same region as the box in (A) but with the RecB and DNA components shown and details of the interactions. (C) The same site but for Gam and RecB. Note that there is some mimicry of the hydrophobic stacking interactions in both complexes.
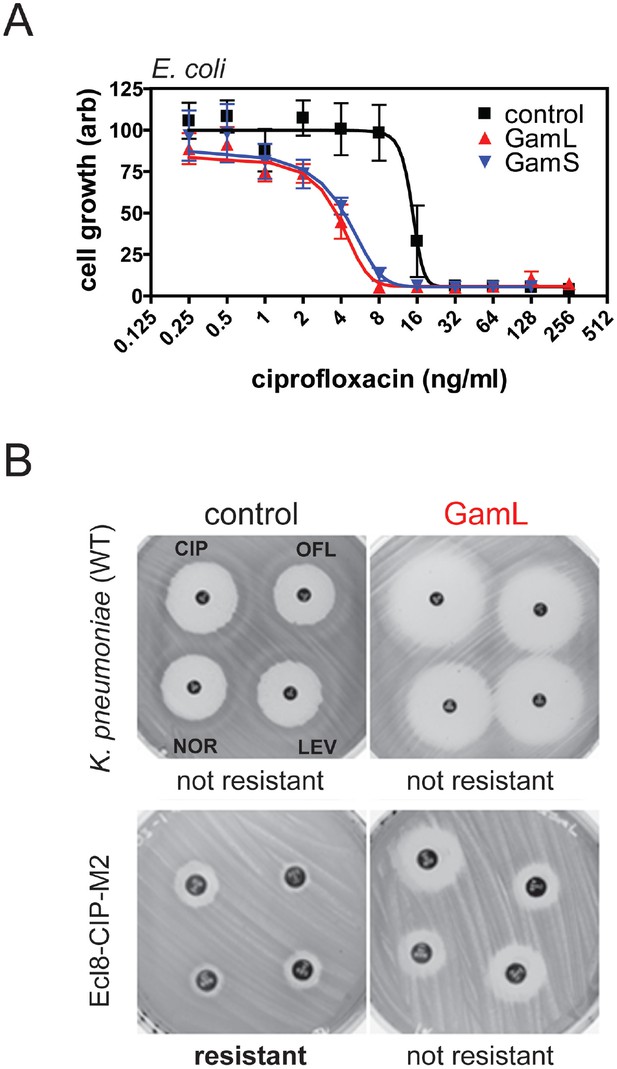
Inhibition of bacterial DSB repair potentiates fluoroquinolone antibiotics.
(A) Ciprofloxacin minimum inhibitory concentration (MIC) assay against E. coli MG1655 cells in the presence or absence of Gam isoforms as indicated. Experiments were performed as described in the Materials and methods in the presence of arabinose to induce expression of the small or large isoforms of Gam. Control experiments were performed under identical conditions with the empty pBADK expression vector. (B) Disc susceptibility assays are standardised tests that quantify antibacterial susceptibility in terms of an inhibition zone diameter, and also classify bacterial strains as either resistant, or not resistant, to antibacterial agents based on expected drug efficacy in a clinical setting. The experiments were performed and interpreted in accordance with CLSI guidelines (CLSI, 2015) with a range of fluoroquinolones (CIP; ciprofloxacin, OFL; ofloxacin, NOR; norfloxacin, LEV, levoxofloxacin). Illustrative results for wild type K. pneumoniae (Ecl8) and a resistant strain (Ecl8-CIP-M2) are shown using the GamL isoform. Note that the magnification of the plates is different for the two strains as is apparent from the disc sizes. The inhibition zone diameters for all strains, with both GamL and GamS, are summarised in Table 1.
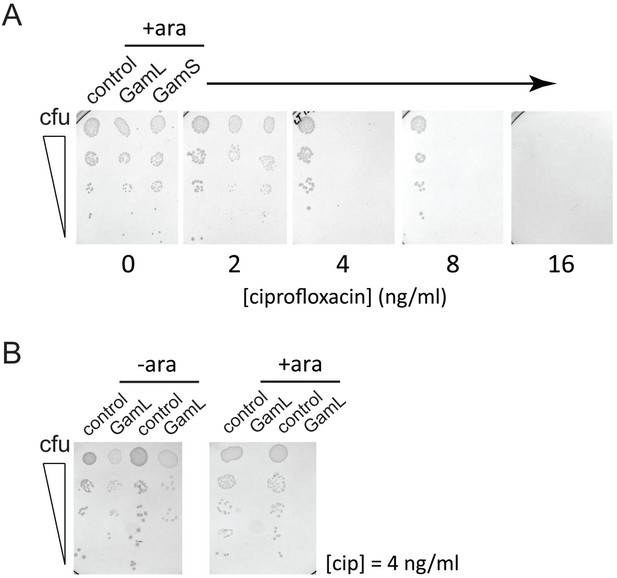
Spot tests for ciprofloxacin sensitivity on agar plates.
(A) Spot tests show the viability of E.coli MG1655 at varying ciprofloxacin concentrations and in the presence of pBADK vectors expressing no protein (control), GamL or GamS as indicated and in the presence of 1% (w/v) arabinose. (B) The ciprofloxacin potentiation effect of Gam is arabinose-dependent. Experiments were performed at 4 ng/ml ciprofloxacin. Spot tests were performed by making five-fold serial dilutions of a ~105 colony forming units (cfu)/ml starting culture of E. coli MG1655 and spot plating onto agar supplemented with kanamycin (50 ug/ml), arabinose (±1% w/v as indicated) and ciprofloxacin (as indicated).
Videos
Electron density map.
A region of the electron density is shown in the location of the N-terminal helix of the Gam subunit that is located in the ssDNA binding site of the RecB subunit. Side chain density is clearly visible.
Overlay of the DNA substrate and the GamS dimer bound to RecBCD.
https://doi.org/10.7554/eLife.22963.007Details of the interactions between the N-terminal helix of Gam and the RecB subunit.
https://doi.org/10.7554/eLife.22963.011Tables
Disc susceptibility assay for K. pneumoniae Ecl8wt, derived mutants and clinical isolates expressing GamL or GamS from pBADK. The disc susceptibility assay were performed according to the CLSI protocol (CLSI, 2015) using Mueller-Hinton agar with 0.2% (w/v) arabinose to stimulate expression of cloned genes (in bracket) and 30 mg/L kanamycin to select for the pBADK plasmids. Resistance breakpoints are as set by the CLSI (CLSI, 2015). Values shaded designate resistance. Values reported are the means of three repetitions rounded to the nearest integer.
K. pneumoniae strains | Diameter of growth inhibition zone (mm) around fluoroquinolone disc | |||
|---|---|---|---|---|
Fluoroquinolone (µg in disc) | ||||
Ciprofloxacin (5) | Levofloxacin (5) | Norfloxacin (10) | Ofloxacin (5) | |
Ecl8wt pBADK (Control) | 34 | 32 | 30 | 30 |
Ecl8wt pBADK (GamL) | 40 | 37 | 37 | 37 |
Ecl8wt pBADK (GamS) | 46 | 42 | 41 | 39 |
Ecl8-CIP- M1 pBADK (Control) | 24 | 21 | 20 | 19 |
Ecl8-CIP-M1 pBADK (GamL) | 31 | 27 | 24 | 24 |
Ecl8-CIP-M1 pBADK (GamS) | 28 | 27 | 25 | 25 |
Ecl8-CIP-M2 pBADK (Control) | 14 | 12 | 11 | 8 |
Ecl8-CIP-M2 pBADK (GamL) | 21 | 17 | 16 | 14 |
Ecl8-CIP-M2 pBADK (GamS) | 21 | 18 | 16 | 14 |
R16 pBADK (Control) | 8 | 8 | 8 | 6 |
R16 pBADK (GamL) | 17 | 16 | 16 | 16 |
R16 pBADK (GamS) | 15 | 15 | 15 | 12 |
R20 pBADK (Control) | 15 | 13 | 14 | 11 |
R20 pBADK (GamL) | 20 | 19 | 19 | 15 |
R20 pBADK (GamS) | 18 | 17 | 17 | 15 |





