COPI mediates recycling of an exocytic SNARE by recognition of a ubiquitin sorting signal
Figures
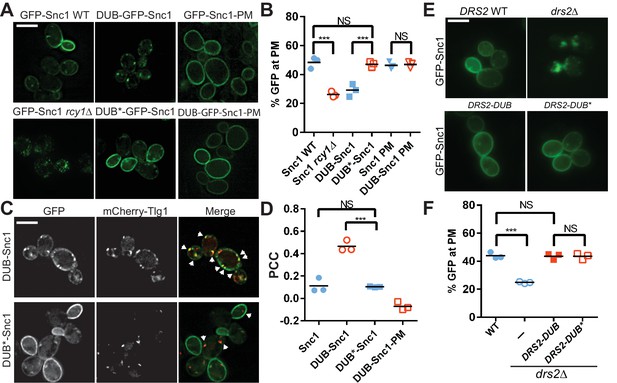
Ubiquitination is required for Snc1 recycling.
(A) Fusion of catalytically active deubiquitinase (DUB), but not the inactive form (DUB*) to GFP-Snc1 blocks its recycling comparably to rcy1∆. Mutation of an endocytic signal (PM) in Snc1 prevents accumulation of DUB-GFP-Snc1-PM in cytosolic punctae. (B) Quantification of GFP intensity at the plasma membrane. At least 50 cells for three biological replicates of each genotype were analyzed, and the value and mean for each biological replicate were plotted. (C) DUB-GFP-Snc1 accumulates in punctae marked by mCherry-Tlg1. The arrowheads highlighted the punctae showing colocalized GFP-Snc1 with mCherry-Tlg1. (D) Pearson correlation coefficient (PCC) GFP-Snc1 with mCherry-Tlg1. Each biological replicate plotted includes at least 20 cells. (E) Fusion of DUB to Drs2 does not disrupt the ability of Drs2 to support Snc1 recycling. (F) Quantification of GFP intensity at the plasma membrane for the cells in (E). Each biological replicate includes at least 50 cells. Statistical differences in (B), (D) and (E) were determined using a one-way ANOVA on the means of three biological replicates (***p<0.001; NS, p>0.05). Scale bar in (A), (C) and (E) represents 5 µm.
-
Figure 1—source data 1
This spreadsheet contains the three means of GFP intensity at the plasma membrane data used to generate the dot plots shown in Figure 1B and F, and the three means of Pearson correlation coefficient data used to generate the dot plots shown in Figure 1D.
Statistical analysis data used to compare the significant difference has been included.
- https://doi.org/10.7554/eLife.28342.003
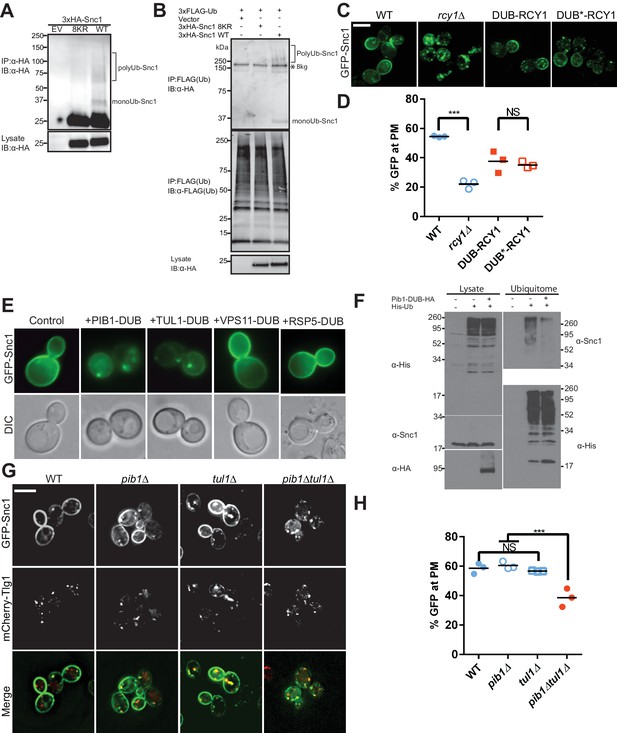
Snc1 is extensively modified with polyUb chains.
(A) Proteins were immunoprecipitated from strains expression an empty vector, a lysineless 3XHA-Snc1-8KR, or WT 3XHA-Snc1 with anti-HA beads, and then immunoblotted using the anti-HA antibody. Monoubiquitinated and polyubiquitinated Snc1 forms are indicated (based on predicted mobilities). (B) Ubiquitinated proteins were immunoprecipitated using anti-FLAG antibodies from strains expressing 3X-FLAG-Ub and either an empty vector, a lysineless 3XHA-Snc1-8KR, or WT 3XHA-Snc1, and then immunoblotted for HA-Snc1 or FLAG-Ub. Monoubiquitinated and polyubiquitinated Snc1 forms are indicated (based on predicted mobilities). The lysine-less 3XHA-Snc1-8KR is a specificity control and the asterisk indicates background bands. (C) Rcy1 appears to play a role in Snc1 recycling that is independent of Ub ligase activity. A fusion of DUB or DUB* to the amino terminus of Rcy1 caused a partial defect in Snc1 recycling when expressed in rcy1∆ cells (BY4742 YJL204C). However, there was no significant difference between DUB and DUB*, indicating that the effect of the DUB is unrelated to its deubiquitinase activity. (D) Quantification of GFP intensity at the plasma membrane. (E) WT cells (BY4742) overexpressing GFP-Snc1 and DUB fusions with several candidate E3 Ub ligases. PIB1-DUB and TUL1-DUB were the only ligase-DUB fusions that caused a GFP-Snc1 recycling defect. (F) DUB tagged Pib1 significantly reduced endogenous polyubiquitinated Snc1. Ubiquitinated proteins (Ubiquitome) were recovered from cells expressing His-tagged Ub with or without Pib1-DUB and probed for endogenous, untagged Snc1 and His-Ub. Much less polyubiquitinated Snc1 was recovered from the ubiquitome in cells expressing DUB-Pib1. DUB-Pib1-HA expression was confirmed by immunoblot with anti-HA antibody. (G) The pib1∆ (PLY5293) and tul1∆ (PLY5294) single mutants recycled GFP-Snc1 normally, but the pib1∆ tul1∆ (PXY64) double mutant displayed a recycling defect. (H) Quantification of GFP intensity at the plasma membrane for cells shown in (G). Each biological replicate includes at least 50 cells for data plotted in (D) and (H). Statistical differences were determined using a one-way ANOVA on the means of three biological replicates. (***p<0.001; NS, p>0.05). Scale bar in (C) and (G) represents 5 µm.
-
Figure 2—source data 1
This spreadsheet contains the three means of GFP intensity at the plasma membrane of Rcy1 mutant cells and pib1, tul1 mutant cells used to generate the dot plots shown in Figure 1D and H.
Statistical analysis data used to determine significant differences has been included.
- https://doi.org/10.7554/eLife.28342.007
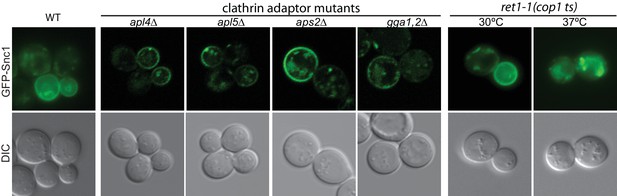
Inactivation of a COPI temperature-sensitive allele (ret1-1) at the non-permissive temperature (37°C) blocked GFP-Snc1 recycling.
By contrast, GFP-Snc1 still could localize to the plasma membrane in the clathrin adaptor AP1 (apl4Δ), AP3 (apl5Δ), AP2 (aps2Δ) or GGA (gga1,2Δ) mutants.
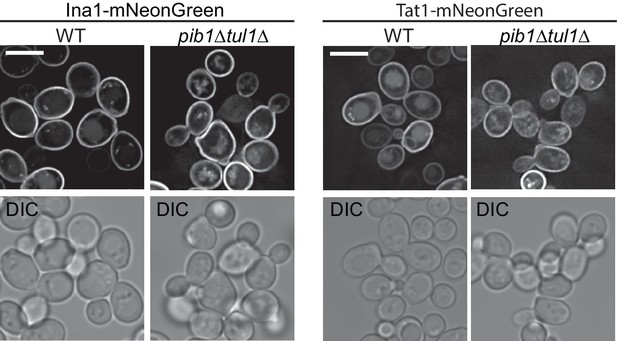
Plasma membrane proteins Ina1 and Tat1 were tagged with mNeonGreen and visualized in WT (wild-type) and pib1∆ tul1∆ double mutant cells by fluorescence microscopy.
Vacuoles are more fragmented in the pib1∆ tul1∆ strain, but no difference was noted in the marker distribution between internal and plasma membrane pools. Scale bar represents 5 µm.
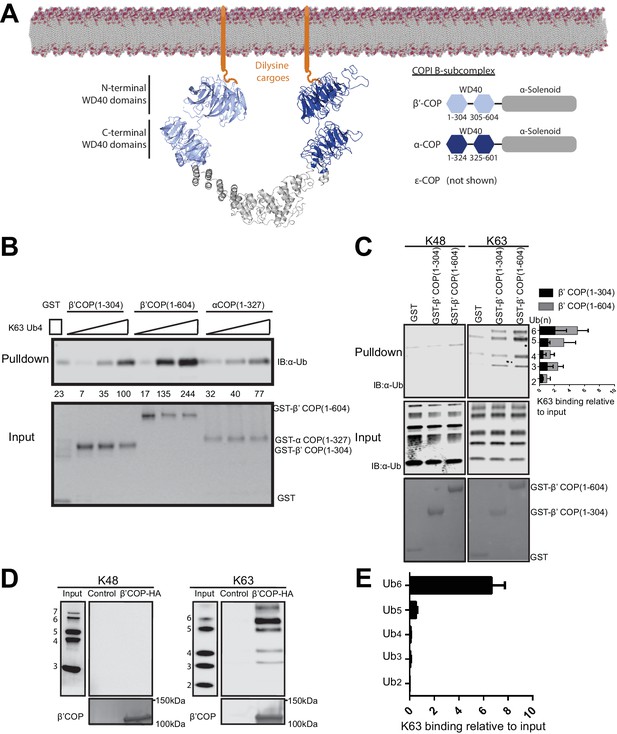
WD40 repeat propeller domains of COPI bind K63-linked polyubiquitin.
(A) Structures of α- and β'-COP from the COPI B-subcomplex shown binding dilysine cargo in the membrane. (B) GST-β'COP (1-604), GST-β'COP (1-304) and GST-αCOP (1-327) bind K63-linked tetraUb relative to the GST only control. 0.5 μM of GST and GST tagged WD40 proteins immobilized glutathione beads were incubated 125 nM, 250 nM or 500 nM of K63-Ub4. Values are tetraUb band intensities from this experiment. (C) Both GST-β'COP (1-604) and GST-β'COP (1-304) preferentially binds long K63-linked chains of Ub. Quantification of K63 binding relative to input (100*(band signal intensity – corresponding GST lane)/input band intensity). The values represent mean ± SEM from three independent binding experiments. (D) COPI isolated from yeast on anti-HA beads also preferentially binds long K63-linked polyUb, but not K48-linked Ub. (E) Quantification of K63-linked Ub polymers binding relative to input. The values represent mean ± SEM from three independent binding experiments (100*band signal intensity/input band intensity).
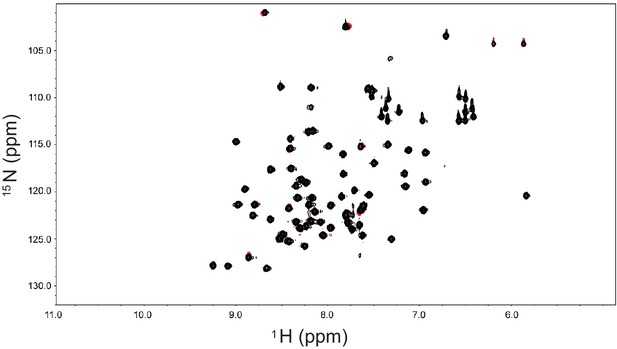
The N-terminal propeller of β'-COP (1-304) does not bind significantly to monoUb.
HSQC spectra of 125 µM 15N-labeled ubiquitin in 40 mM NaPO4 (pH = 7.1) in the absence and presence of 1.25 mM unlabeled N-terminal propeller of β’COP. Spectra show an almost complete overlay of 15N Ub alone (red) and in the presence of a 10-fold excess of the N-terminal propeller of β'-COP (black).
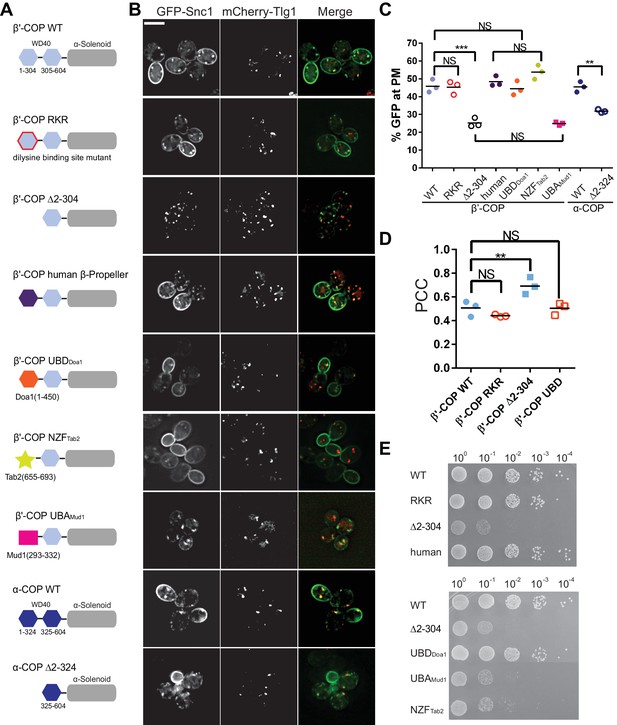
Ub binding by β'-COP is required to sort GFP-Snc1 to the plasma membrane.
(A and B) Deletion of the N-terminal WD40 propeller from β'-COP (∆2–304) disrupts recycling of GFP-Snc1 but mutation of residues within this propeller required for dilysine motif binding (RKR) have no effect. Replacement of the N-terminal propeller with a linkage independent ubiquitin-binding domain (UBD) from Doa1, a K63-specific Npl4 Zinc Finger (NZF) domain from Tab2, or the N-terminal propeller from human β'-COP restored Snc1 recycling. In contrast, the K48-linkage specific ubiquitin pathway associated (UBA) domain from Mud1 failed to restore GFP-Snc1 recycling. Deletion of N-terminal WD40 propeller of α-COP caused a partial Snc1 recycling defect. Scale bar, 5 µm. (C) Quantification of GFP intensity at the plasma membrane. Each biological replicate includes at least 50 cells and individual biological replicates value and mean are shown. Statistical differences were determined using a one-way ANOVA on the means of the three biological replicates (***p<0.001; NS, p>0.05). (D) Quantification analysis of colocalization between GFP-Snc1 and mCherry-Tlg1 in WT and β'-COP mutant cells using Pearson correlation coefficient. Each replicate includes at least 20 cells and individual biological replicates value and mean were shown. (E) Serial dilution growth assay of β'-COP mutants. The β'-COP dilysine motif binding mutant (RKR) had no effect on growth, but deletion of the first propeller (∆2–304) caused slow growth. Replacement of the first propeller with the UBD or the human N-terminal propeller, but not NZF or UBA domains, restored WT growth. One of four replicates is shown.
-
Figure 4—source data 1
This spreadsheet contains the GFP intensity at the plasma membrane of COPI mutants used to generate the dot plots shown in Figure 4C, and the means of Pearson correlation coefficient data of COPI mutants used to generate the dot plots shown in Figure 4D.
Statistical analysis data used to determine significant differences has been included.
- https://doi.org/10.7554/eLife.28342.012
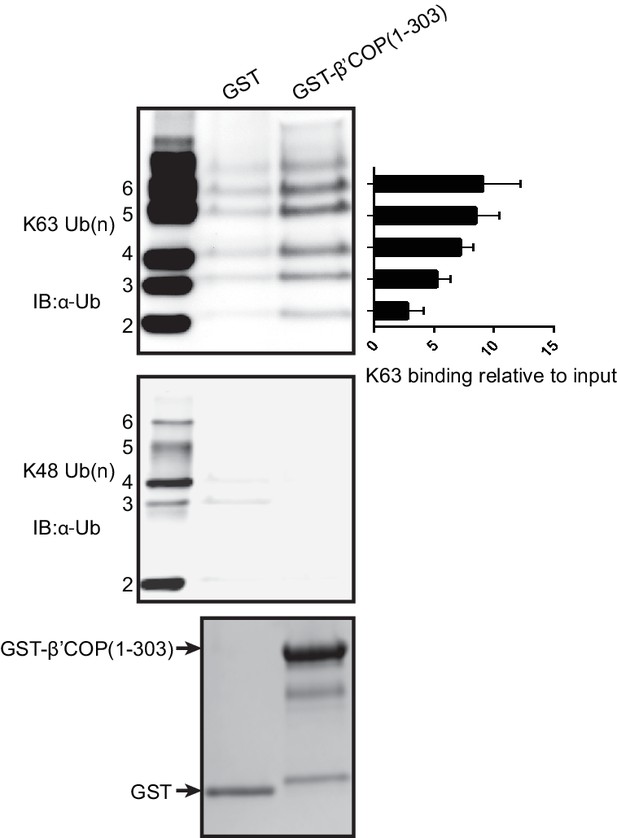
GST tagged human β'-COP (1-303) preferentially binds long K63-linked (upper panel) but not K48-linked (middle panel) chains of Ub.
Bait proteins GST and GST-β'COP (1-303) eluted from the glutathione beads were stained with Coomassie (bottom panel). The graph displays quantification of K63-Ub binding relative to input (100*(band signal intensity – corresponding GST lane)/input band intensity). The values represent mean ± SEM from three independent binding experiments.
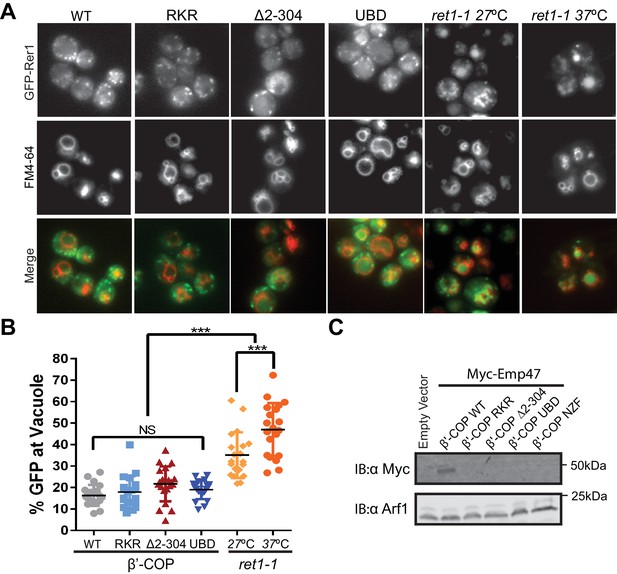
β'-COP interaction with ubiquitin has no role in the COPI-dependent trafficking of Rer1 or Emp47 at the Golgi complex.
(A) Deletion of N-terminal WD40 propeller of β'-COP does not alter the retrograde trafficking of Rer1 from cis-Golgi to endoplasmic reticulum. The indicated β’ COP mutant cells expressing GFP-Rer1 were labeled with 2 nM FM4-64 for 20 min at 30 ˚C then chased for 2 hr to label vacuole membranes. An α-COP temperature-sensitive mutant (ret1-1) expressing GFP-Rer1 was labeled with 2 nM FM4-64 at 27 ˚C for 20 min, then chased at 27 ˚C or 37 ˚C for 2 hr. (B) Quantification of GFP-Rer1 in the vacuole. The percentage values of GFP intensity in the vacuole for 20 cells were plotted. The mean and standard deviation were shown. Statistical differences were determined using a one-way ANOVA on the means of three biological replicates. (***p<0.001; NS, p>0.05). (C) Immunoblot showing that Myc-Emp47, an early Golgi COPI cargo, is missorted into the vacuole and degraded in strains expressing β'-COP RKR or ∆2–304, but stability is not restored when the N-terminal propeller is replaced with Ub binding domains. Immunoblot using anti-Arf1 is used as the loading control.
-
Figure 5—source data 1
This spreadsheet contains the percentage of GFP intensity at the vacuole labeled with FM4-64 for individual β’-COPI mutants cells and ret1-1 mutants cells used to generate the dot plots shown in Figure 5B.
Statistical analysis data used to determine significant differences has been included.
- https://doi.org/10.7554/eLife.28342.014
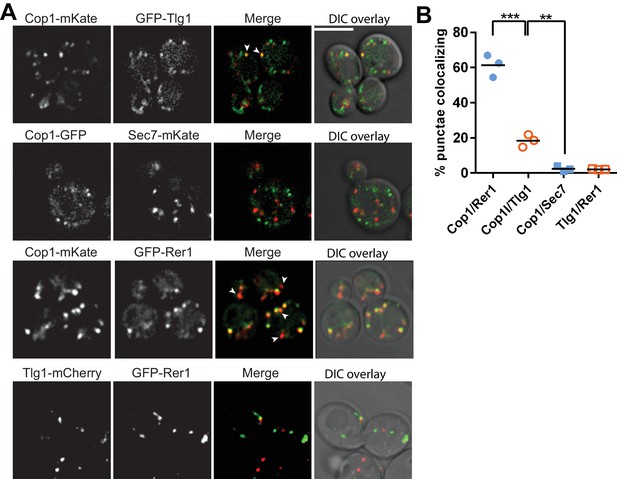
A small pool of COPI co-localizes with Tlg1 in yeast.
(A and B) Co-localization of Cop1 (α-COP) with markers for the early endosome/TGN (Tlg1), early Golgi (Rer1), or TGN (Sec7). Tlg1 was also co-localized relative to Rer1 to make sure there was no significant overlap between these early Golgi and early endosome/TGN markers. Scale bar, 5 µm. Statistical differences were determined using a one-way ANOVA on the means of three biological replicates (***p<0.001; **p<0.01; NS, p>0.05).
-
Figure 6—source data 1
This spreadsheet contains three means of percentage for Cop1 with Rer1, Tlg1 or Sec7 and Tlg1 with Sec7 data used to generate the dot plots shown in Figure 6B.
Statistical analysis data used to determine significant differences has been included.
- https://doi.org/10.7554/eLife.28342.016
Additional files
-
Supplementary file 1
List of plasmids used in this study.
- https://doi.org/10.7554/eLife.28342.017
-
Supplementary file 2
List of strains used in this study.
- https://doi.org/10.7554/eLife.28342.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28342.019





