In vivo experiments do not support the charge zipper model for Tat translocase assembly
Figures
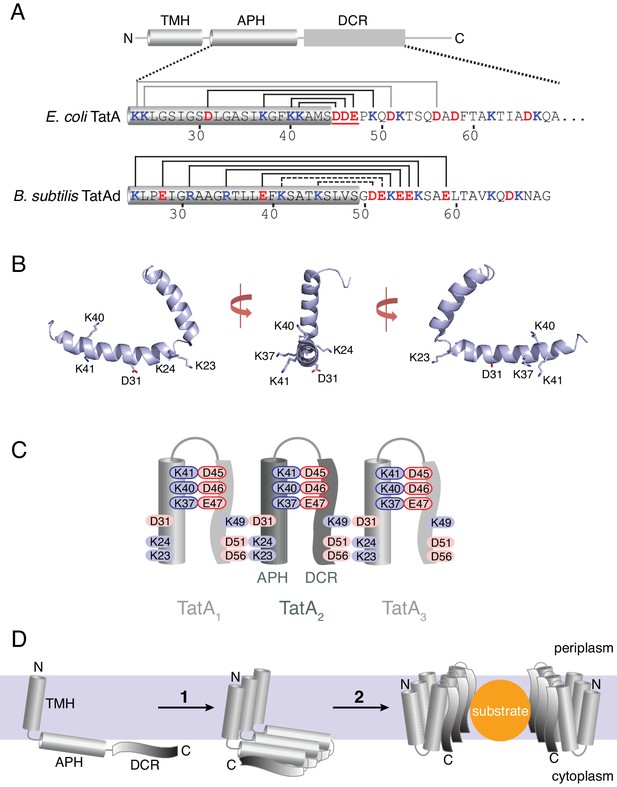
The charge zipper model for TatA assembly.
(A) Cartoon showing the TatA domain structure, comprising a transmembrane helix (TMH), an amphipathic helix (APH), and a densely-charged region (DCR). Below the cartoon are shown the amino acid sequences for E. coli TatA (top) and B. subtilis TatAd (bottom) starting at the beginning of the APH and with acidic (red) and basic (blue) residues indicated. The APH is assigned in each case from the corresponding NMR structures (PDB 2MN7 and 2L16) and is indicated by a gray cylinder. The E. coli TatA sequence is C-terminally truncated to residue 69. Salt bridge pairs predicted by Walther and co-workers are indicated above each sequence. For E. coli TatA the predicted salt bridge pairs tested in this study are indicated in black and the acidic DDE motif targeted in this study is underlined. For B. subtilis TatAd, the assigned intra- and inter-molecular pairs are distinguished using dotted or solid lines respectively. (B) Structure of E. coli TatA (PDB 2MN7) shown in three orientations with the charged APH side chains indicated. (C) Schematic diagram of the charge zipper model for TatA folding and assembly applied to E. coli TatA. Folding back the DCR against the APH is proposed to allow pairing of amino acids with complementary charges to form either intra-molecular or inter-molecular salt bridges. Three adjacent TatA protomers are shown with the residues of the acidic DDE motif and their potential salt bridge partners outlined in red and blue respectively. The charge zipper model does not predict which residue pairs would form inter- and intra-molecular salt bridges and one of several possible configurations is represented here. (D) The salt bridges shown in (C) are proposed to mediate self-assembly of multiple adjacent TatA molecules (1). The assembled APH/DCR units are then proposed to insert across the membrane to form the passage for substrate protein transport (2).
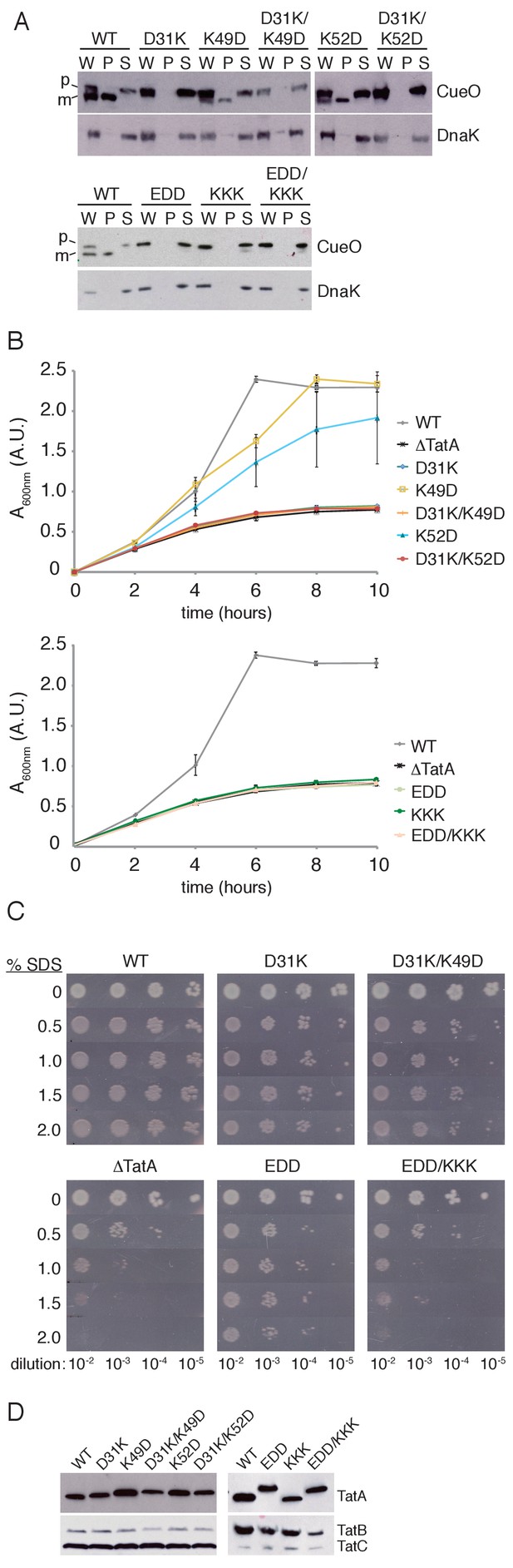
Transport activity of TatA charge zipper variants.
All TatA variants were expressed from the phage lambda attachment site in the ΔtatA ΔtatE strain JARV16. Mutant strains are identified by the amino acid substitutions present in TatA where EDD is K37E/K40D/K41D, and KKK is D45K/D46K/E47K. WT refers to the ΔtatE strain J1M1, and ΔTatA refers to the parental strain JARV16. (A) Whole cell (W), periplasm (P), and spheroplast (S) fractions of cells overproducing CueO from plasmid pQE80-CueO were subject to immunoblotting with antibodies against CueO or the cytoplasmic marker protein DnaK. m is the transported form of CueO from which the signal peptide has been removed and p the precursor protein. (B) Growth of the strains when cultured in LB/glycerol/TMAO medium under anoxic conditions. Error bars represent the S.E.M of three biological replicates. (C) Serial ten-fold dilutions of log-phase cultures were spotted onto LB-agar containing the indicated amount of SDS. (D) Immunoblot of membranes isolated from the strains used in A-C, probed with TatA antibodies (upper panels) or antibodies against TatB and TatC (lower panels).
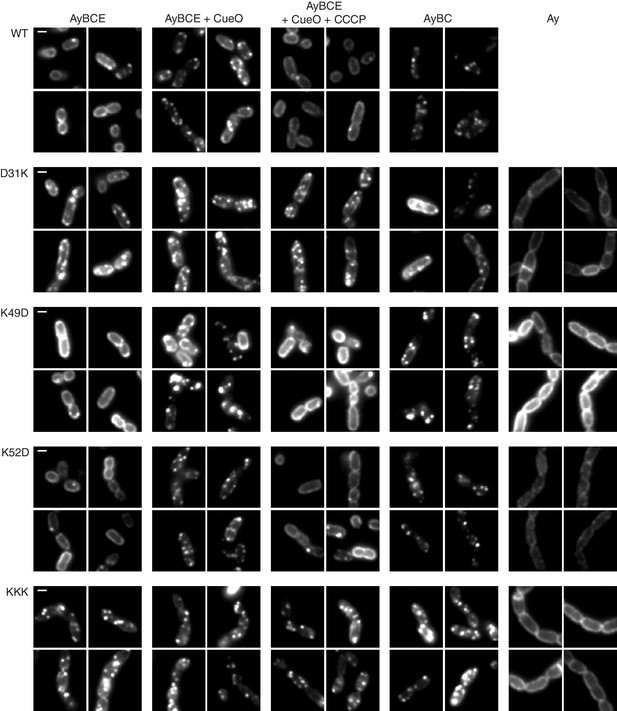
TatA oligomerization behavior of charge zipper variants.
Representative fluorescence images of TatA-YFP in E. coli cells. A tatA-yfp fusion was expressed from the chromosome in three different backgrounds: ELV16 λAry which contains all other tat genes (designated AyBCE), JARV16 λAry which lacks tatE (designated AyBC), or DADE λAry which possesses no other tat genes (designated Ay). The TatA-YFP variant produced is indicated to the left of the panels, where WT is the parental protein and KKK is a D45K/D46K/E47K variant. Where indicated, CueO was overproduced from plasmid pQE80-CueO by adding IPTG to early exponential phase cultures for 30 min prior to imaging (+CueO columns). 50 μM CCCP was subsequently added, as indicated (+CCCP column), and the cells incubated for 30–45 min prior to imaging. Scale bar = 1 μm.
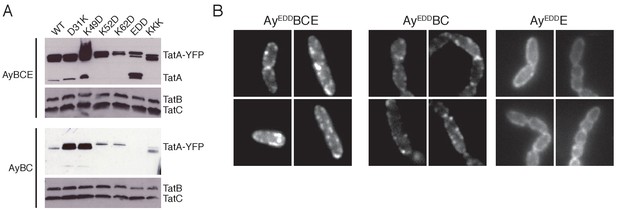
TatA oligomerization behavior of additional charge zipper variants.
(A) Immunoblot of membranes isolated from the strains used in panel (B) and in Figure 3, probed with TatA antibodies (upper panels) or antibodies against TatB and TatC (lower panels). (B) Representative fluorescence images of strains ELV16 λAry EDD (AyEDDBCE), JARV16 λAry EDD (AyEDDBC), and MΔABC λAry EDD (AyEDDE). The scaling used to display these images differs from that employed in Figure 3 and uses 1000 a.u. as the minimum (black) and 4000 a.u. as the maximum (white).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (Escherichia coli) | MC4100 | Casadaban and Cohen, 1979 | ||
| strain, strain background (E. coli) | MC1061 | Casadaban and Cohen, 1980 | ||
| strain, strain background (E. coli) | J1M1 | Sargent et al., 1998 | MC4100 ΔtatE | |
| strain, strain background (E. coli) | JARV16 | Sargent et al., 1999 | MC4100 ΔtatA ΔtatE | |
| strain, strain background (E. coli) | ELV16 | Sargent et al., 1999 | MC4100 ΔtatA | |
| strain, strain background (E. coli) | DADE | Wexler et al., 2000 | MC4100 ΔtatABCD ΔtatE | |
| strain, strain background (E. coli) | MΔABC | Alcock et al., 2013 | MC4100 ΔtatABC::apra | |
| strain, strain background (E. coli) | JARV16 λA D31K | This paper | JARV16 attB::PtatAtatAD31K (kanr) | |
| strain, strain background (E. coli) | JARV16 λA K49D | This paper | JARV16 attB::PtatAtatAK49D (kanr) | |
| strain, strain background (E. coli) | JARV16 λA D31K/K49D | This paper | JARV16 attB::PtatAtatAD31K,K49D (kanr) | |
| strain, strain background (E. coli) | JARV16 λA K52D | This paper | JARV16 attB::PtatAtatAK52D (kanr) | |
| strain, strain background (E. coli) | JARV16 λA D31K/K52D | This paper | JARV16 attB::PtatAtatAD31K,K52D (kanr) | |
| strain, strain background (E. coli) | JARV16 λA EDD | This paper | JARV16 attB::PtatAtatAK37E,K40D,K41D (kanr) | |
| strain, strain background (E. coli) | JARV16 λA KKK | This paper | JARV16 attB::PtatAtatAD45K,D46K,E47K (kanr) | |
| strain, strain background (E. coli) | JARV16 λA EDD/KKK | This paper | JARV16 attB::PtatAtatAK37E,K40D,K41D,D45K,D46K,E47K (kanr) | |
| strain, strain background (E. coli) | JARV16 λAry | This paper | JARV16 attB::PtatAtatA-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | JARV16 λAry D31K | This paper | JARV16 attB::PtatAtatAD31K-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | JARV16 λAry K49D | This paper | JARV16 attB::PtatAtatAK49D-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | JARV16 λAry K52D | This paper | JARV16 attB::PtatAtatAK52D-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | JARV16 λAry EDD | This paper | JARV16 attB::PtatAtatA K37E,K40D,K41D-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | JARV16 λAry KKK | This paper | JARV16 attB::PtatAtatA D45K,D46K,E47K-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | ELV16 λAry D31K | This paper | ELV16 attB::PtatAtatAD31K-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | ELV16 λAry K49D | This paper | ELV16 attB::PtatAtatAK49D-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | ELV16 λAry K52D | This paper | ELV16 attB::PtatAtatAK52D-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | ELV16 λAry EDD | This paper | ELV16 attB::PtatAtatA K37E,K40D,K41D- EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | ELV16 λAry KKK | This paper | ELV16 attB::PtatAtatA D45K,D46K,E47K-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | DADE λAry D31K | This paper | DADE attB::PtatAtatAD31K-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | DADE λAry K49D | This paper | DADE attB::PtatAtatAK49D-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | DADE λAry K52D | This paper | DADE attB::PtatAtatAK52D-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | DADE λAry KKK | This paper | DADE attB::PtatAtatA D45K,D46K,E47K-EAK-eyfpA206K (kanr) | |
| strain, strain background (E. coli) | MΔABC λAry EDD | This paper | MC4100 ΔtatABC::apra attB::PtatAtatA K37E,K40D,K41D-EAK-eyfpA206K (kanr) | |
| antibody | anti-CueO | This paper | Rabbit poly clonal against CueO mature domain. Affinity purified (1:200) | |
| antibody | anti-DnaK | Abcam | Abcam ab69617; Clone 8E2/2 | Mouse monoclonal (1:20000) |
| antibody | anti-TatA | Alcock et al., 2016 | Rabbit polyclonal against TatA soluble domain (1:5000) | |
| antibody | anti-TatB | Alcock et al., 2016 | Rabbit polyclonal against TatB C-terminal peptide. Affinity purified (1:400) | |
| antibody | anti-TatC | Alcock et al., 2016 | Rabbit polyclonal against TatC C-terminal peptide. Affinity purified (1:1000) | |
| recombinant DNA reagent | pQE80-CueO | Leake et al., 2008 | Synthesis of E. coli CueO with a C-terminal his6 tag | |
| recombinant DNA reagent | pKSUniA | Koch et al., 2012 | pBluescript-based vector carrying PtatA-tatA | |
| recombinant DNA reagent | pBSTatAry | Alcock et al., 2013 | pBluescript-based vector carrying PtatA-tatA-EAK-eyfpA206K | |
| recombinant DNA reagent | pRS552 | Simons et al., 1987 | Shuttle vector for integration of DNA at the E. coli phage lambda attachment site (attB) | |
| recombinant DNA reagent | λRS45 | Simons et al., 1987 | Phage for integration of DNA at the E. coli phage lambda attachment site (attB) | |
| sequence-based reagent | TatA D31K F | Sigma-Aldrich, St. Louis, Missouri | Oligonucleotide GGCTCCATCGGTTCCAAACTTGGTGCGTCGATC | |
| sequence-based reagent | TatA D31K R | Sigma-Aldrich | Oligonucleotide GATCGACGCACCAAGTTTGGAACCGATGGAGCC | |
| sequence-based reagent | TatA K49D F | Sigma-Aldrich | Oligonucleotide CAATGAGCGATGATGAACCAGATCAGGATAAAACCAGTCAGG | |
| sequence-based reagent | TatA K49D R | Sigma-Aldrich | Oligonucleotide CCTGACTGGTTTTATCCTGATCTGGTTCATCATCGCTCATTG | |
| sequence-based reagent | TatA K52D F | Sigma-Aldrich | Oligonucleotide GAACCAAAGCAGGATGATACCAGTCAGGATGCTG | |
| sequence-based reagent | TatA K52D R | Sigma-Aldrich | Oligonucleotide CAGCATCCTGACTGGTATCATCCTGCTTTGGTTC | |
| sequence-based reagent | TatA EDD F | Sigma-Aldrich | Oligonucleotide CTTGGTGCGTCGATCGAAGGCTTTGATGATGCAATGAGCGATGATG | |
| sequence-based reagent | TatA EDD R | Sigma-Aldrich | Oligonucleotide CATCATCGCTCATTGCATCATCAAAGCCTTCGATCGACGCACCAAG | |
| sequence-based reagent | TatA KKK F | Sigma-Aldrich | Oligonucleotide GCTTTAAAAAAGCAATGAGCAAAAAGAAACCAAAGCAGGATAAAACC | |
| sequence-based reagent | TatA KKK R | Sigma-Aldrich | Oligonucleotide GGTTTTATCCTGCTTTGGTTTCTTTTTGCTCATTGCTTTTTTAAAGC | |
| sequence-based reagent | TatA zip2 F | Sigma-Aldrich | Oligonucleotide CTTGGTGCGTCGATCGAAGGCTTTGATGATGCAATGAGCAAAAAG | |
| sequence-based reagent | TatA zip2 R | Sigma-Aldrich | Oligonucleotide CTTTTTGCTCATTGCATCATCAAAGCCTTCGATCGACGCACCAAG |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30127.006





