Tropical Disease: Dissecting the schistosome cloak
Schistosomiasis is a devastating disease that affects around 200 million people worldwide. It is caused by parasitic flatworms known as flukes or schistosomes, which can infect people through exposure to contaminated drinking water and poor sanitation. Current treatments only work after infection has occurred, which makes it difficult to eradicate this disease completely (World Health Organization, 2018).
Schistosomes belong to the animal clade called neodermata, which consists of approximately 100,000 species of parasitic flatworms (Littlewood, 2006). The key evolutionary innovation defining this clade is a skin-like structure called the tegument that allows the parasites to withstand particularly harsh environments, such as the human digestive system and the bloodstream. It also helps the worms to absorb nutrients and attach to their hosts, important adaptions that potentially enabled the worms to become parasites.
Since all parasitic flatworms have a tegument, it is a prime target for drug development. Indeed, the only drug that is currently available for the treatment of schistosomiasis, praziquantel, is thought to work by dissolving the tegument, although the mechanisms involved remain unknown (Chai, 2013). We also do not fully understand how adult schistosomes generate and maintain their teguments.
Some studies indicate that schistosomes can survive inside human hosts for decades without getting detected by the immune system (Basch, 1991). Until recently, visualizing the tegument has required the use of electron microscopy, a technique that is difficult to combine with other strategies for highlighting cells. Now, in eLife, James Collins and colleagues at the University of Texas Southwestern Medical Center (UTSW), James Cook University and the Wellcome Trust Sanger Institute – including George Wendt of UTSW as first author – report a straightforward method to label the tegument with fluorescent dyes (Wendt et al., 2018).
Soaking mature schistosomes in water causes the tegument to swell, which damages its membrane and so allows the fluorescent dyes to seep into it. Unlike electron microscopy, fluorescent dyes are compatible with other cell biology techniques, such as lineage-tracing, in situ hybridization and antibody labeling. By using these methods, Wendt et al. were able to determine the identity and birthdate of the cells in the tegument. The results showed that far from being a static structure, the tegument constantly incorporates new cells, which are produced by adult stem cells (Figure 1; Collins et al., 2013). The rapid turnover of cells in the tegument resembles the production of epithelial cells in free-living flatworms called planarians, from which parasitic flatworms have evolved (Laumer et al., 2015; Egger et al., 2015).
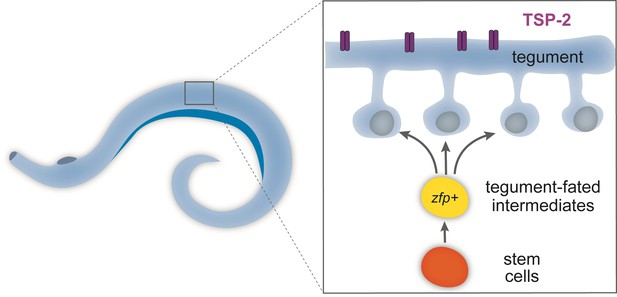
Schematic of the tegument in adult flatworms.
Parasitic flatworms (schistosomes) live in the human bloodstream and are covered by a skin-like structure called the tegument, which is constantly replenished by newly produced stem cells (see inset) and so protects the worms from being detected by the host immune system. The tegument (blue) is made of cells that have fused together to form a continuous structure and express a protein called TSP-2 (purple). The cells in the tegument derive from stem cells (orange). These stem cells divide to produce intermediate cells (yellow), which mature to produce the cells (blue) that fuse to form the tegument. These tegument-fated intermediate cells express ‘zinc-finger’ proteins (zfp) and are necessary to build the skin-like structure. Creating drugs that could block these genes may present a new opportunity to treat schistosomiasis.
Wendt et al. then used an antibody that recognizes TSP-2: this protein, which is found on the surface of the tegument, is a molecular target in efforts to develop a schistosome vaccine (Pearson et al., 2012). This antibody allowed the researchers to purify the tegument cells, to determine the genes that are expressed by these cells, and to identify the proteins that are required to build the tegument. Wendt et al. identified two ‘zinc-finger’ proteins, which are only found in flatworms. These two proteins are necessary for forming the cells that produce the tegument. When one of these proteins is blocked, the schistosomes can no longer adhere to substrates, which may affect their ability to interact with the host.
Intriguingly, free-living planarians also rely on stem cells that express a similar zinc-finger protein to generate their epidermis (van Wolfswinkel et al., 2014; Tu et al., 2015). So, even though they have very different surfaces, parasitic flatworms and free-living flatworms employ strikingly similar molecular processes to make their ‘skin’. These conserved mechanisms may therefore be a prime target for drugs that treat schistosomiasis by preventing the formation of the tegument.
The dynamic nature of the tegument demonstrated in the study of Wendt et al. raises key questions about schistosomes and their host-parasite relationships. In particular, it is unclear how the integrity of the tegument can be maintained while its cells are constantly turning over. Over the course of its life, a schistosome displays antigens on its exterior that, presumably, reveal to its host that it is a foreign invader. So how does the worm regulate which proteins remain on its surface when new cells constantly join the tegument?
It has previously been hypothesized that rapid shedding of the tegument could be a mechanism to evade the immune system of the host, to potentially prevent antigens from being exposed (Abath and Werkhauser, 1996). More research is needed to determine how the constant replacement of the tegument cells contributes to the schistosome’s ability to evade host immune systems. Furthermore, expanding the repertoire of potential treatments remains a high priority, given the extreme number and diversity of parasitic flatworms, and the debilitating effects of the diseases they cause in livestock and in humans.
References
-
BookSchistosomes: Development, Reproduction, and Host RelationsOxford University Press.
-
Praziquantel treatment in trematode and cestode infections: an updateInfection & Chemotherapy 45:32–43.https://doi.org/10.3947/ic.2013.45.1.32
-
BookThe evolution of parasitism in flatwormsIn: Maule A. G, Marks N. J, editors. Parasitic Flatworms: Molecular Biology, Biochemistry, Immunology and Physiology. Wallingford: CABI. pp. 1–36.
-
Enhanced protective efficacy of a chimeric form of the schistosomiasis vaccine antigen Sm-TSP-2PLoS Neglected Tropical Diseases 6:e1564.https://doi.org/10.1371/journal.pntd.0001564
Article and author information
Author details
Publication history
- Version of Record published: April 27, 2018 (version 1)
Copyright
© 2018, Adler
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 999
- views
-
- 60
- downloads
-
- 4
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Developmental Biology
Inhibitory G alpha (GNAI or Gαi) proteins are critical for the polarized morphogenesis of sensory hair cells and for hearing. The extent and nature of their actual contributions remains unclear, however, as previous studies did not investigate all GNAI proteins and included non-physiological approaches. Pertussis toxin can downregulate functionally redundant GNAI1, GNAI2, GNAI3, and GNAO proteins, but may also induce unrelated defects. Here, we directly and systematically determine the role(s) of each individual GNAI protein in mouse auditory hair cells. GNAI2 and GNAI3 are similarly polarized at the hair cell apex with their binding partner G protein signaling modulator 2 (GPSM2), whereas GNAI1 and GNAO are not detected. In Gnai3 mutants, GNAI2 progressively fails to fully occupy the sub-cellular compartments where GNAI3 is missing. In contrast, GNAI3 can fully compensate for the loss of GNAI2 and is essential for hair bundle morphogenesis and auditory function. Simultaneous inactivation of Gnai2 and Gnai3 recapitulates for the first time two distinct types of defects only observed so far with pertussis toxin: (1) a delay or failure of the basal body to migrate off-center in prospective hair cells, and (2) a reversal in the orientation of some hair cell types. We conclude that GNAI proteins are critical for hair cells to break planar symmetry and to orient properly before GNAI2/3 regulate hair bundle morphogenesis with GPSM2.
-
- Computational and Systems Biology
- Developmental Biology
Organisms utilize gene regulatory networks (GRN) to make fate decisions, but the regulatory mechanisms of transcription factors (TF) in GRNs are exceedingly intricate. A longstanding question in this field is how these tangled interactions synergistically contribute to decision-making procedures. To comprehensively understand the role of regulatory logic in cell fate decisions, we constructed a logic-incorporated GRN model and examined its behavior under two distinct driving forces (noise-driven and signal-driven). Under the noise-driven mode, we distilled the relationship among fate bias, regulatory logic, and noise profile. Under the signal-driven mode, we bridged regulatory logic and progression-accuracy trade-off, and uncovered distinctive trajectories of reprogramming influenced by logic motifs. In differentiation, we characterized a special logic-dependent priming stage by the solution landscape. Finally, we applied our findings to decipher three biological instances: hematopoiesis, embryogenesis, and trans-differentiation. Orthogonal to the classical analysis of expression profile, we harnessed noise patterns to construct the GRN corresponding to fate transition. Our work presents a generalizable framework for top-down fate-decision studies and a practical approach to the taxonomy of cell fate decisions.






