Bone-Targeting Drugs: From vesicle to cytosol
Despite its appearance, bone is a highly metabolic and dynamic tissue that is composed of a vast network of cells called osteocytes that are embedded in a matrix made mostly of collagen and various salts of calcium and phosphate. These osteocytes sense regions of damaged or weakened bone, and 'instruct' bone-destroying cells (called osteoclasts) and bone-forming cells (osteoblasts) to, respectively, remove old bone and deposit new bone (Figure 1). Hence, like a team of road-repairers, the osteocytes, osteoclasts and osteoblasts work together to repair bone and maintain our skeleton in good health (Crockett et al., 2011).
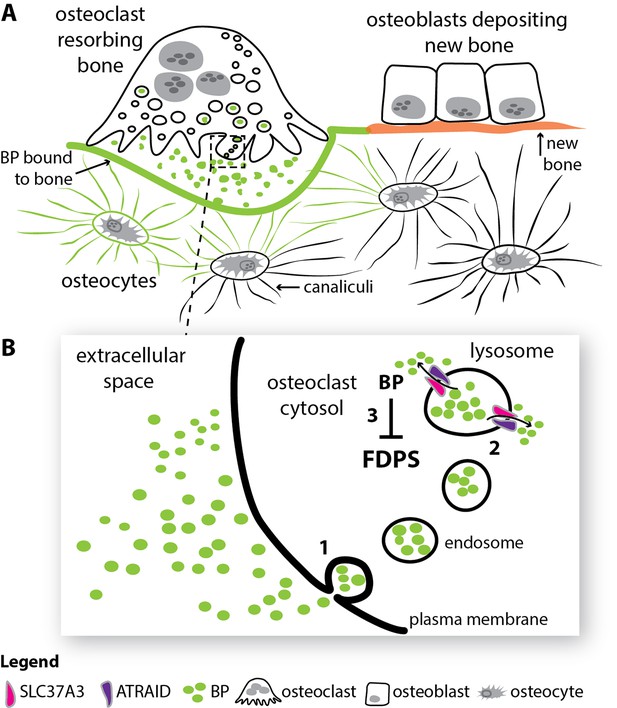
How bisphosphonates act in bone.
(A) Old and damaged bone is constantly being broken down by cells called osteoclasts in a process called resorption (top left), while new bone is deposited by cells called osteoblasts (top right). Cells called osteocytes (bottom) influence both of these processes through a spidery system of tiny canals called canaliculi. After entering the circulation, the drug bisphosphonate (green) binds very effectively to calcium ions on the bone mineral surface. During resorption, the bisphosphonate on the bone surface is released into the acidic extracellular space beneath the osteoclast. (B) The bisphosphonate in the extracellular space is engulfed into osteoclasts via a process called endocytosis (1). The resulting endosomes mature to form structures called lysosomes, and two proteins, SLC37A3 and ATRAID, then interact in the membrane of the lysosome to allow the bisphosphonate to enter the cytosol (2). Once in the cytosol, the nitrogen-containing bisphosphonates inhibit an enzyme called FDPS and prevent the osteoclast from breaking down bone (3). BP: bisphosphonate; FDPS: farnesyl diphosphate synthase; SLC37A3: solute carrier family 37 member A3.
In young adult life there is usually a balance between the amount of old bone broken down and the amount of new bone formed by this repair process, so there is no net gain or loss of bone mass. However, in diseases that affect the skeleton, such as post-menopausal osteoporosis or cancers growing in bone, this delicate balance can be disturbed by osteoclasts being over-active, which leads to excessive bone destruction and fractures. Drugs called bisphosphonates – which inhibit osteoclasts – have been used for more than three decades to treat such diseases and protect the skeleton from potentially catastrophic bone loss, although researchers still do not fully understand how they work. Now, in eLife, Erin O'Shea of Harvard University and the Howard Hughes Medical Institute (HHMI) and colleagues – including Zhou Yu as first author – report the answer to one of the remaining questions about these drugs (Yu et al., 2018).
Bisphosphonates are synthetic molecules that closely resemble the chemical structure of pyrophosphate, which is a natural by-product of numerous metabolic reactions. Importantly, bisphosphonates have two negatively-charged phosphonate groups that enable them to bind calcium ions very effectively, and hence to localize rapidly to any exposed calcium on the bone surface (Rogers et al., 2011). The mechanisms used by bisphosphonates to inhibit osteoclasts remained a mystery for several decades after they were first used in the clinic, but this did not stop the development of improved versions of the drugs (Russell et al., 2008). Eventually it was discovered that nitrogen-containing bisphosphonates (N-BPs), which are now widely used to treat osteoporosis and other bone diseases, work by inhibiting an enzyme called FDPS inside the osteoclasts (Luckman et al., 1998; Bergstrom et al., 2000; Dunford et al., 2001). The N-BP molecules displace the lipid substrates that the FDPS enzyme usually acts on, locking the enzyme in an inactive state (Kavanagh et al., 2006; Rondeau et al., 2006). Without FDPS activity, osteoclasts are no longer able to degrade bone (Rogers et al., 2011).
However, one question remained: how do the N-BPs and other bisphosphonates actually reach the FDPS enzyme, which is in the cytosol of the osteoclasts? There was little or no evidence that a receptor on the plasma membrane was involved (Thompson et al., 2006). Studies with fluorescently-tagged bisphosphonates showed that they first entered the osteoclasts via endocytosis – a process that involves the cell membrane folding inwards and then pinching off to create a vesicle inside the cell (Coxon et al., 2008). But how do the drugs leave these vesicles – which are enclosed by a membrane – to enter the cytosol? Bisphosphonate molecules have a large negative charge, which rules out passive diffusion across the vesicle membrane, which in turn suggests the possibility of a hitherto unidentified transport mechanism (Thompson et al., 2006).
Yu et al. – who are based at Harvard, HHMI, UCSF, MIT, the Broad, Koch and Whitehead Institutes, and Washington University – report that they have identified a protein called SLC37A3 that is required for the release of N-BP molecules from vesicles into the cytosol. Using a CRISPR-based approach to screen for genes that, when missing, confer resistance to bisphosphonates, they identified SLC37A3 as the gene with the strongest effect. Although the exact function of the SLC37A3 protein remains to be clarified, related members of this protein family are involved in the transport of charged molecules across membranes (Cappello et al., 2018).
Yu et al. found that SLC37A3 interacts and co-localizes with a protein called ATRAID at the vesicle membrane (Figure 1). Importantly, vesicles isolated from cells that did not express SLC37A3 or ATRAID appeared unable to release N-BP molecules, and these cells were much less sensitive to the pharmacological effect of N-BPs.
This new transport mechanism identified by Yu et al. raises interesting questions about how the SLC37A3/ATRAID complex specifically recognizes N-BP molecules, and how it transports them across the membrane of the vesicle. It will also be worthwhile to determine whether differences in the expression of SLC37A3 or ATRAID account for the different sensitivity of osteoclasts and other cell types to N-BP molecules, or whether variants in these genes affect the clinical responsiveness of patients to these drugs. Nevertheless, these elegant studies explain how negatively-charged N-BP molecules can gain access to the cell cytosol after endocytosis and, as a result, go on to benefit huge numbers of patients with potentially devastating bone diseases.
References
-
Alendronate is a specific, nanomolar inhibitor of farnesyl diphosphate synthaseArchives of Biochemistry and Biophysics 373:231–241.https://doi.org/10.1006/abbi.1999.1502
-
Bone remodelling at a glanceJournal of Cell Science 124:991–998.https://doi.org/10.1242/jcs.063032
-
Structure-activity relationships for inhibition of farnesyl diphosphate synthase in vitro and inhibition of bone resorption in vivo by nitrogen-containing bisphosphonatesJournal of Pharmacology and Experimental Therapeutics 296:235–242.
Article and author information
Author details
Publication history
- Version of Record published: June 27, 2018 (version 1)
Copyright
© 2018, Rogers et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,199
- views
-
- 133
- downloads
-
- 6
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Biochemistry and Chemical Biology
- Cell Biology
Mediator of ERBB2-driven Cell Motility 1 (MEMO1) is an evolutionary conserved protein implicated in many biological processes; however, its primary molecular function remains unknown. Importantly, MEMO1 is overexpressed in many types of cancer and was shown to modulate breast cancer metastasis through altered cell motility. To better understand the function of MEMO1 in cancer cells, we analyzed genetic interactions of MEMO1 using gene essentiality data from 1028 cancer cell lines and found multiple iron-related genes exhibiting genetic relationships with MEMO1. We experimentally confirmed several interactions between MEMO1 and iron-related proteins in living cells, most notably, transferrin receptor 2 (TFR2), mitoferrin-2 (SLC25A28), and the global iron response regulator IRP1 (ACO1). These interactions indicate that cells with high MEMO1 expression levels are hypersensitive to the disruptions in iron distribution. Our data also indicate that MEMO1 is involved in ferroptosis and is linked to iron supply to mitochondria. We have found that purified MEMO1 binds iron with high affinity under redox conditions mimicking intracellular environment and solved MEMO1 structures in complex with iron and copper. Our work reveals that the iron coordination mode in MEMO1 is very similar to that of iron-containing extradiol dioxygenases, which also display a similar structural fold. We conclude that MEMO1 is an iron-binding protein that modulates iron homeostasis in cancer cells.
-
- Biochemistry and Chemical Biology
- Structural Biology and Molecular Biophysics
NADPH oxidases (NOX) are transmembrane proteins, widely spread in eukaryotes and prokaryotes, that produce reactive oxygen species (ROS). Eukaryotes use the ROS products for innate immune defense and signaling in critical (patho)physiological processes. Despite the recent structures of human NOX isoforms, the activation of electron transfer remains incompletely understood. SpNOX, a homolog from Streptococcus pneumoniae, can serves as a robust model for exploring electron transfers in the NOX family thanks to its constitutive activity. Crystal structures of SpNOX full-length and dehydrogenase (DH) domain constructs are revealed here. The isolated DH domain acts as a flavin reductase, and both constructs use either NADPH or NADH as substrate. Our findings suggest that hydride transfer from NAD(P)H to FAD is the rate-limiting step in electron transfer. We identify significance of F397 in nicotinamide access to flavin isoalloxazine and confirm flavin binding contributions from both DH and Transmembrane (TM) domains. Comparison with related enzymes suggests that distal access to heme may influence the final electron acceptor, while the relative position of DH and TM does not necessarily correlate with activity, contrary to previous suggestions. It rather suggests requirement of an internal rearrangement, within the DH domain, to switch from a resting to an active state. Thus, SpNOX appears to be a good model of active NOX2, which allows us to propose an explanation for NOX2’s requirement for activation.






