Sexual Differentiation: A matter of timing
The fate of a cell during development depends on several factors, such as its location in the body, developmental stage, and sex. However, most cells in the body do not ‘know’ what sex they are and function the same way, even though male and female cells have different sex chromosomes. How, then, do sexually dimorphic cells – the cells that are responsible for differences between males and females, such as differences in the adult brain – learn their sex during development?
The sex of an animal can modify the development of cells, such as neurons, in several ways. For example, males and females can produce the same type of cells, but then cause targeted cell death in a subset of cells in one of the sexes (Figure 1A; Kimura et al., 2008; Sanders and Arbeitman, 2008). Alternatively, an animal’s sex can modify the number of cell divisions, producing extra cells in one sex (Figure 1B; Emmons, 2018; Taylor and Truman, 1992; Sanders and Arbeitman, 2008). Or, cells that are identical at first can be modified in different ways in males and females (Figure 1C). This mechanism is common in the worm Caenorhabditis elegans (reviewed in Portman, 2017), but less common in insects like the fruit fly (see, for example, Kohl et al., 2013).
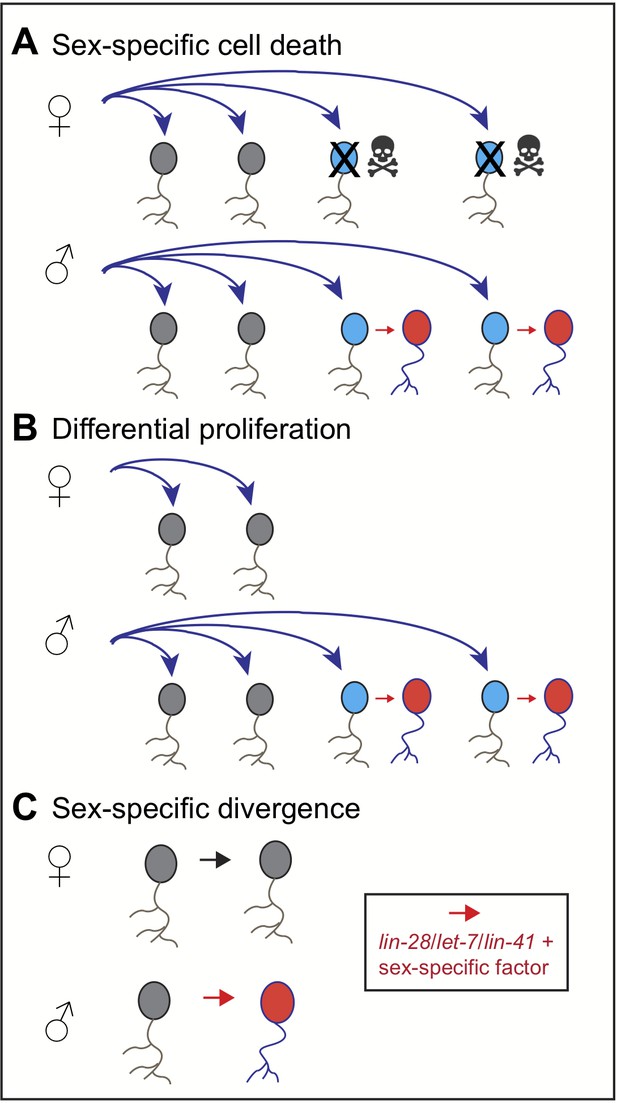
Sexual dimorphism in neurons.
The sex of an animal can modify the development of sex-specific neurons in several ways. (A) Male and female animals may develop the same type of neurons (shown in grey), but over the course of development, one sex (females in this case) may lose some of the neurons (blue) due to programmed cell death (skulls). The neurons of the other sex (male neurons in this case) then acquire sex-specific features (red). (B) Input from the sex-specific pathway can modify the number of cell divisions, producing extra neurons (blue) in one sex, which then acquire their sex-specific features (red). (C) Alternatively, neurons can develop sex-specific features that affect their shape and/or properties (red). In C. elegans, this is mediated by combination of the heterochronic pathway (which involves the genes let-7, lin-28 and lin-41) and sex-specific genes, such as lin29a or homologs of doublesex.
In C. elegans, most of these differences arise during the fourth stage of larval development, just before they become sexually active, but how do cells know when to become different? Now, in eLife, Oliver Hobert of Columbia University and colleagues – including Laura Pereira as first author – report that the heterochronic pathway (which regulates the larval development of worms) is also involved in controlling the timing of sexual differentiation in the nervous system of C. elegans (Pereira et al., 2019).
Pereira et al. – who are based at Columbia, Rochester, and Basel – reveal that three genes (let-7, lin-28 and lin-41) control when sexual maturation takes place in the neurons of C. elegans. Mutation of let-7 precociously initiates sex-specific changes in neurons, while overexpression overrides these changes. Furthermore, in young worms, lin-41 represses the production of a newly identified version of a gene called lin-29a, which is sexually specific. As the worms mature, however, the protein of lin-28 is lost, which allows let-7 to deactivate lin-41. In turn, lin-29a is then expressed in a subset of neurons. These neurons then turn on male-specific genes and adopt a male-specific shape and function (in the manner shown in Figure 1C). Both let-7 and lin-28 have been shown to control the timing of sexual differentiation in mice and humans, providing an intriguing hint of deeply conserved mechanisms (see, for example, Corre et al., 2016; Zhu et al., 2010; Chen et al., 2017).
The researchers discovered that the activation of lin-29a controls the sex-specific features of a neuron, called the AIM interneuron, which is important for the behavior of males. Male worms that lack this gene move in a way that is typical for hermaphrodites (modified females that can self-fertilize), providing a convincing demonstration that lin-29a provides the sex-specific input in the regulation of male genes. Pereira et al. further found that the pathway that determines the sex of a cell, including the gene tra-1, also regulates the production of lin-29a.
In other cells in C. elegans, homologs of a gene called doublesex, rather than lin29a, shape the sex-specific traits. The gene doublesex is well-known for being involved in the sex-specific development of the nervous system in insects, although another gene called fruitless (which is not present in worms), is even more important in this process. Given the similarities, it is tempting to draw parallels between lin-29a and fruitless, though no direct homology has been identified.
It is unknown whether a heterochronic pathway similar to the one in C. elegans affects sex-specific cell fate decisions in fruit flies. The genes fruitless and doublesex only start to exhibit sex-specific expression in the nervous system at the end of the last larval stage and the early pupal stages (but as soon as the neurons have formed), which can then result in sexual dimorphism (as in Figure 1). This is different from what Pereira et al. report in C. elegans, where the pathway acts on many types of neurons at roughly the same stage, often well after they have been specified. Whether any neurons in Drosophila exhibit a similar delay in sex-specific fate specification after patterning is unknown.
The links to let-7 and lin-28 in the sexual differentiation of vertebrates raise the question of how precisely these genes are affecting their neural development during puberty; whether the number or types of neurons are modified by this pathway, as they are in C. elegans; and whether similar connections to homologs of lin-29a or doublesex-like genes exist. The study of Pereira et al. has set the stage for future work in flies and vertebrates to test potentially shared components and interactions.
References
-
Association study of LIN28B in girls with precocious pubertyJournal of Pediatric Endocrinology and Metabolism 30:663–667.https://doi.org/10.1515/jpem-2016-0101
-
Sex-specific regulation of weight and puberty by the Lin28/let-7 axisJournal of Endocrinology 228:179–191.https://doi.org/10.1530/JOE-15-0360
-
Neural circuits of sexual behavior in Caenorhabditis elegansAnnual Review of Neuroscience 41:349–369.https://doi.org/10.1146/annurev-neuro-070815-014056
-
Sexual modulation of sex-shared neurons and circuits in Caenorhabditis elegansJournal of Neuroscience Research 95:527–538.https://doi.org/10.1002/jnr.23912
-
Commitment of abdominal neuroblasts in Drosophila to a male or female fate is dependent on genes of the sex-determining hierarchyDevelopment 114:625–642.
Article and author information
Author details
Publication history
- Version of Record published: January 1, 2019 (version 1)
Copyright
© 2019, Perry and Desplan
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,671
- views
-
- 144
- downloads
-
- 2
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cell Biology
- Neuroscience
Alternative RNA splicing is an essential and dynamic process in neuronal differentiation and synapse maturation, and dysregulation of this process has been associated with neurodegenerative diseases. Recent studies have revealed the importance of RNA-binding proteins in the regulation of neuronal splicing programs. However, the molecular mechanisms involved in the control of these splicing regulators are still unclear. Here, we show that KIS, a kinase upregulated in the developmental brain, imposes a genome-wide alteration in exon usage during neuronal differentiation in mice. KIS contains a protein-recognition domain common to spliceosomal components and phosphorylates PTBP2, counteracting the role of this splicing factor in exon exclusion. At the molecular level, phosphorylation of unstructured domains within PTBP2 causes its dissociation from two co-regulators, Matrin3 and hnRNPM, and hinders the RNA-binding capability of the complex. Furthermore, KIS and PTBP2 display strong and opposing functional interactions in synaptic spine emergence and maturation. Taken together, our data uncover a post-translational control of splicing regulators that link transcriptional and alternative exon usage programs in neuronal development.
-
- Genetics and Genomics
- Neuroscience
Single-cell RNA sequencing reveals the extent to which marmosets carry genetically distinct cells from their siblings.






