IL-21/type I interferon interplay regulates neutrophil-dependent innate immune responses to Staphylococcus aureus
Figures
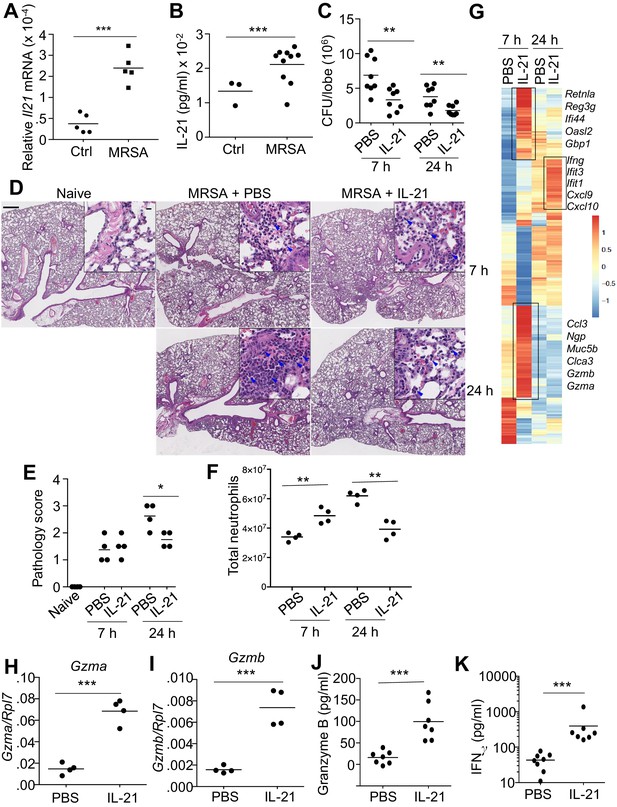
IL-21 enhances killing of pulmonary MRSA in WT mice.
(A, B) Intra-tracheal infection with the USA 300 strain of MRSA induced a significant increase in pulmonary Il21 mRNA (A) and IL-21 protein (B) 24 hr after infection. (C) PBS or 2 μg of IL-21 was administered i.t. to WT mice one day prior to MRSA infection, and lung MRSA CFU were quantitated 7 and 24 hr later. (D, E) Lung immunopathology was assessed in H and E-stained sections of lung tissue from naïve uninfected mice and mice pre-treated with PBS or IL-21 and then infected for 7 or 24 hr with MRSA (in upper left panel, the bar = 500 μm and inset bar = 10 μm) (D), and pathology (E) scores were assessed. (F–K) animals were infected with MRSA as above. (F) Total lung neutrophil cellularity was quantitated by flow cytometry after staining with Ly6G and CD11b. (G) RNA-Seq analysis was performed on total lung tissue mRNA (pools of 5 animals) isolated 7 or 24 hr after treatment of WT mice with PBS or IL-21. Boxed regions include genes mentioned in the text. (H–K) RT-PCR was used to assess expression of Gzma (H) and Gzmb (I) mRNA in lungs treated with IL-21 for 7 hr, and ELISA was used to assess the induction of granzyme B (J) and IFNγ protein (K) in corresponding bronchoalveolar lavage fluid at 7 hr. Data are representative of either three (A, B, C, F, H–K) or two (D, E, G) independent experiments and validation of RNA-Seq was performed by RT-PCR of mRNA from additional mice.
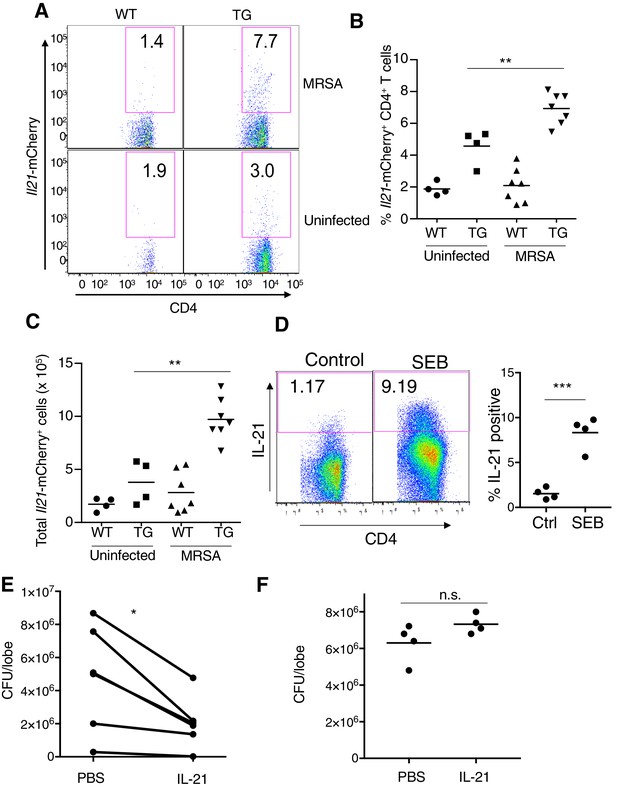
IL-21 is expressed in the lung before and after MRSA infection.
(A–C) IL-21-mCherry transgenic reporter mice were either not infected or infected with MRSA for 24 hr. Lung cells were analyzed by flow cytometry for the percentage of Il21-mCherry+ CD4+ T cells (A, B), and the total number of these cells was also determined (C). The flow plots in (A) are from a representative experiment and (B–C) are pooled from two independent experiments. (D) Human CD4+ T cells were stimulated in vitro without or with SEB and analyzed by flow cytometry for IL-21 expression at 72 hr. Left, flow plot from a representative experiment; right, dot plot summary of 2 independent experiments. (E) Summary of 6 independent experiments in which WT mice were pre-treated with PBS or IL-21, infected with MRSA, and CFU quantitated 7 hr later. (F) Il21r KO mice were treated i.t. with PBS or IL-21, infected with MRSA, and lung CFU quantitated 7 hr later.
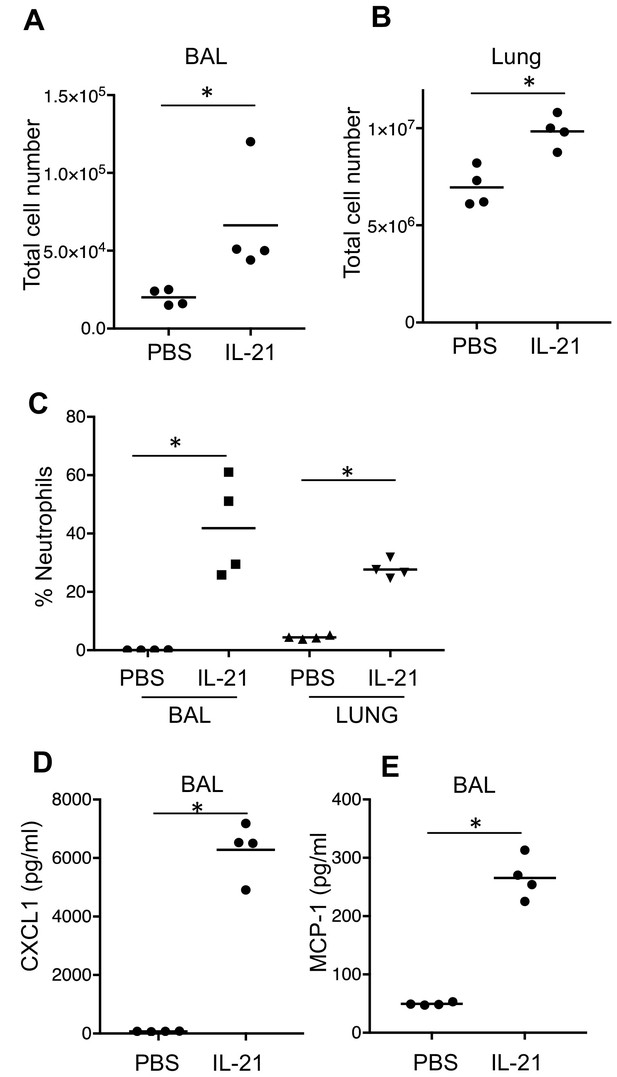
Effects of IL-21 on naïve lung.
(A–B) Total cellularity of lung was assessed in BAL fluid (A) and lung (B) 24 hr after IL-21 intratracheal instillation. (C) Percentage of neutrophils was assessed by flow cytometry at 24 hr after IL-21 treatment. (D–E) Levels of chemokines were measured in BAL fluid at 24 hr after IL-21 treatment.
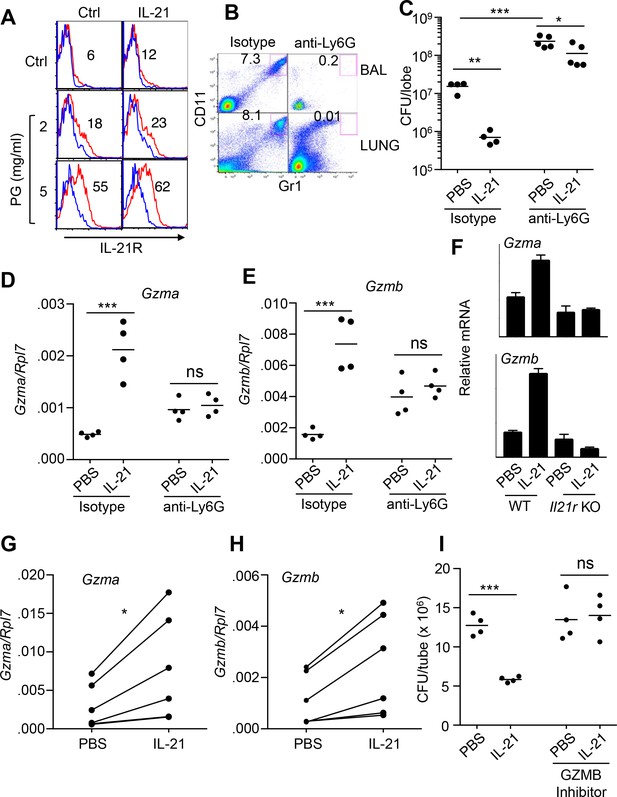
IL-21-mediated granzyme production and MRSA killing by lung neutrophils.
(A) Bone-marrow neutrophils were purified by negative selection using a neutrophil isolation kit (Miltenyi) and either not stimulated or stimulated in vitro with IL-21 (100 ng/ml) for 24 hr in the absence (control) or presence of peptidoglycan (PG) (2 or 5 μg/ml); IL-21R expression on gated Ly6G+CD11b+ neutrophils was detected by flow cytometry (blue = isotype control; red = anti-IL-21R). (B, C) IL-21 lowers the MRSA CFU in a neutrophil-dependent fashion. Mice were treated i.p. with an isotype control mAb or neutrophil-depleted with anti-Ly6G (1A8 mAb), and the efficacy of neutrophil depletion is shown in BAL and lung (B). Mice were then treated with PBS or IL-21 i.t., infected i.t. with MRSA, and CFU quantitated at 24 hr (C). CFU quantitation was performed on the left single lobe, whereas immune populations and RNA were determined using the right lobes. (D, E) Levels of Gzmb and Gzma mRNAs in lung tissue of untreated or neutrophil-depleted mice (treated as in panel C) were quantitated by RT-PCR. (F) Levels of Gzma and Gzmb mRNAs in purified cell-sorted lung neutrophils (>98% pure Ly6G+CD11b+) from either untreated or IL-21-treated WT or Il21r KO mice were measured by RT-PCR and normalized to Rpl7 expression. (G, H) Lung neutrophils were elicited by i.t treatment with heat-killed S. aureus 24 hr prior to isolation, purified, stimulated in vitro for 4 hr with either PBS or IL-21 (100 ng/ml), and Gzma (G) and Gzmb (H) mRNAs assayed by RT-PCR and normalized to Rpl7 expression. (I) Purified HKSA-elicited lung neutrophils were incubated in vitro with MRSA for 3 hr in the presence of PBS or IL-21 either without or with the granzyme B inhibitor, Z-AAD-CMK. MRSA CFU was quantitated by plating serial dilutions on blood agar plates. Representative experiments are shown; each experiment was performed three times with similar results.
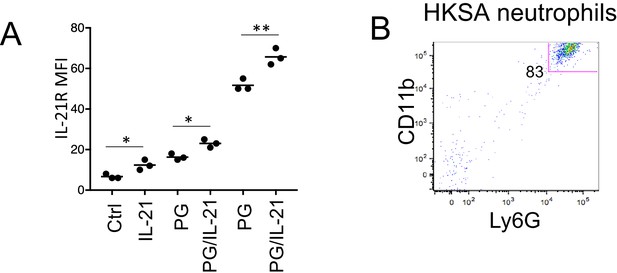
Purity of neutrophil preparations.
(A) Quantitation of IL-21R MFI in neutrophils treated with or without IL-21 in the presence or absence of peptidoglycan (PG). (B) Lung neutrophils from HKSA-stimulated mice were purified via a magnetic neutrophil purification kit (Miltenyi). Shown is CD11b versus Ly6G staining.
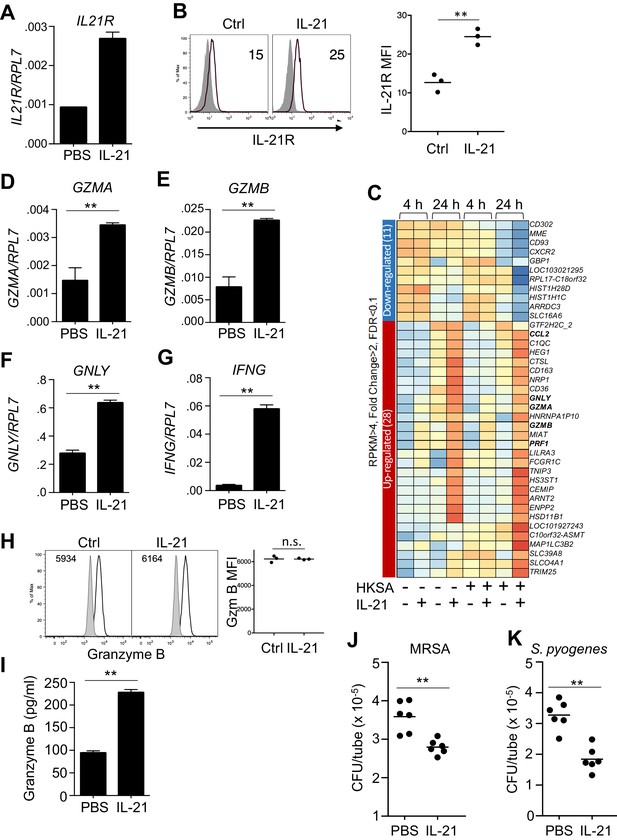
Human neutrophils express IL-21R, and IL-21 induces a program that leads to granzyme B-dependent neutrophil-mediated killing of MRSA.
(A, B) Purified peripheral blood neutrophils were stimulated with PBS (control) or IL-21 (100 ng/ml) for 4 hr, and IL21R mRNA was measured by real-time PCR and normalized to RPL7 expression (A), and IL-21R protein levels were measured by flow cytometry (B); left panel shows isotype control shaded and anti-IL21R black line, with a summary in the right panel). (C) RNA-Seq was performed on neutrophils after 4 or 24 hr incubation with PBS or IL-21 in the absence or presence of heat-killed S. aureus (106/ml). We used HKSA rather than live bacteria in order to allow analysis at 24 hr, as live bacteria would have overgrown the system by then. Genes differentially expressed (fold-change >2.0) are shown. Shown is a representative RNA-Seq analysis. (D – G) Human peripheral blood neutrophils were stimulated for 4 hr in vitro in the presence or absence of IL-21 and GZMA (D), GZMB (E), GNLY (F), and IFNG (G) mRNA levels were quantitated by RT-PCR and normalized to RPL7 expression. (H) Purified human neutrophils were stimulated with IL-21 for 4 hr, fixed, permealized, and stained for intracellular granzyme B protein (gated on CD66b+ cells); MFIs are summarized in right panel. (I) Granzyme B protein was measured by ELISA in supernatants from human peripheral blood neutrophils cultured for 24 hr in either the absence or presence of IL-21 (100 ng/ml). (J, K) Human neutrophils were incubated in vitro with either MRSA (J) or S. pyogenes (K) for 3 hr with PBS or IL-21 (100 ng/ml). MRSA and S. pyogenes CFU were quantitated by plating serial dilutions on blood agar plates. Results shown are representative of 3 independent experiments, except panel C shows one of two similar independent RNA-Seq experiments, each from a different donor.
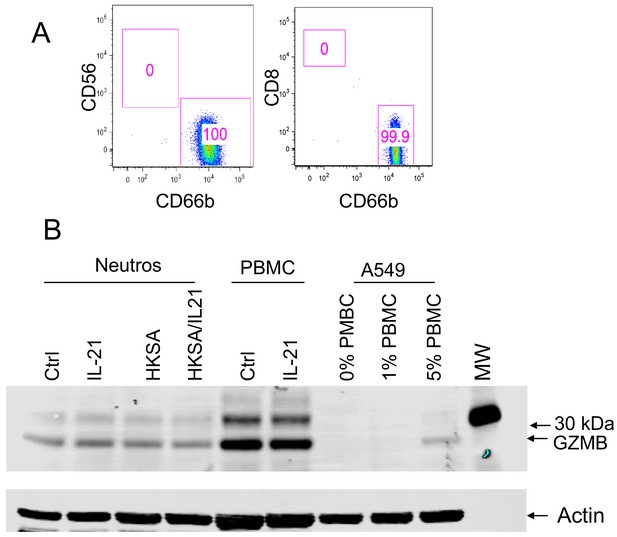
Purity of human neutrophils.
(A) Human peripheral blood neutrophils were purified by a neutrophil negative selection kit (StemCell Technologies). Shown is CD56 versus CD66b and CD8 versus CD66b staining. (B) Western blotting of granzyme B from neutrophils, PBMC, or A549 cells containing 0, 1, or 5% PBMC extracts, confirming that the low level of contamination of purified neutrophils cannot account for the granzyme B expression observed in this population. Also shown is western blotting for actin as a loading control.
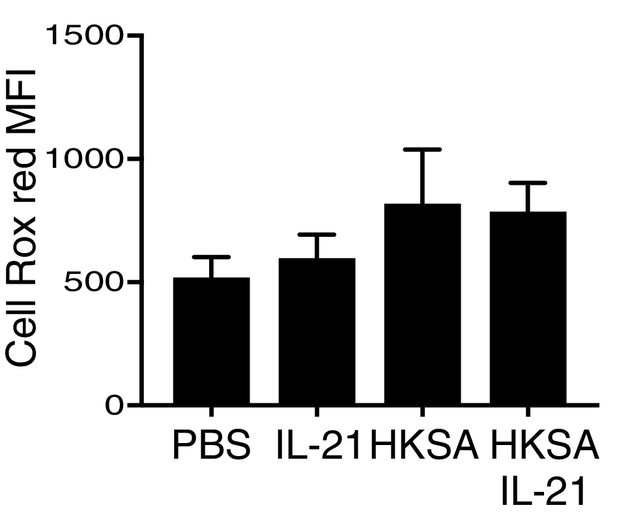
Reactive oxygen species were measured by flow cytometry in CellRox Red loaded peripheral blood neutrophils that were stimulated at 37°C for 30 min with PBS, IL-21, HKSA, or HKSA +IL-21.
https://doi.org/10.7554/eLife.45501.009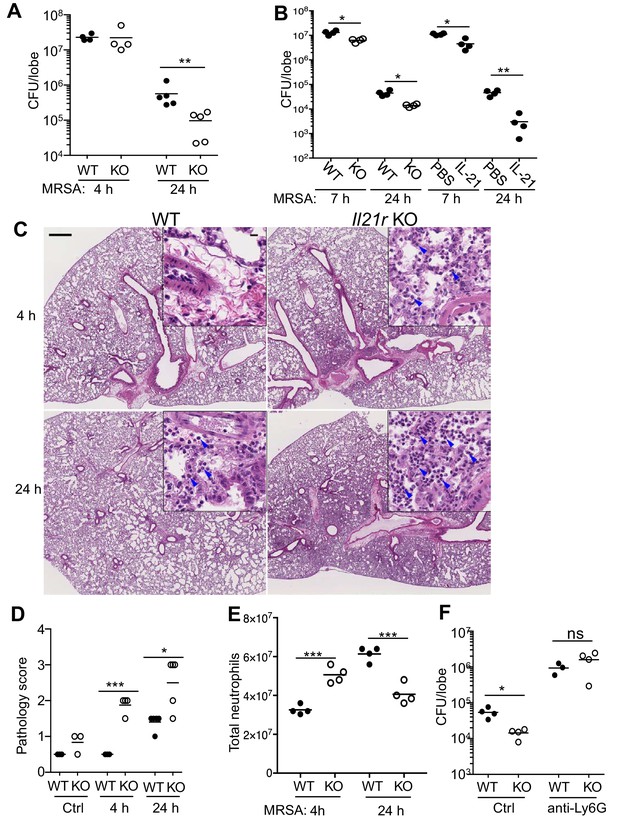
Pulmonary MRSA infection is cleared more efficiently in Il21r KO than in WT mice and this requires neutrophils.
(A) WT and Il21r KO mice were infected intratracheally with MRSA (2 × 107) and CFU in the lung were quantitated at 4 and 24 hr post-infection. (B) WT and Il21r KO mice were infected in parallel with WT mice that had been treated with PBS or 2 μg IL-21, and CFU in the lung were quantitated at 7 and 24 hr post infection. (C–E) Lung immunopathology was assessed in H and E-stained lung sections (bar in upper left panel = 500 μm; bar in inset = 10 μm) (C); lung neutrophil cellularity as assessed by pathology score (D) and flow cytometry (E) were assessed at 4 and 24 hr post-infection. (F) WT or Il21r KO mice were pre-treated with an isotype control antibody or anti-Ly6G to deplete neutrophils, infected with MRSA, and CFU quantitated at 24 hr. Representative experiments are shown; each experiment was performed three times with similar results.
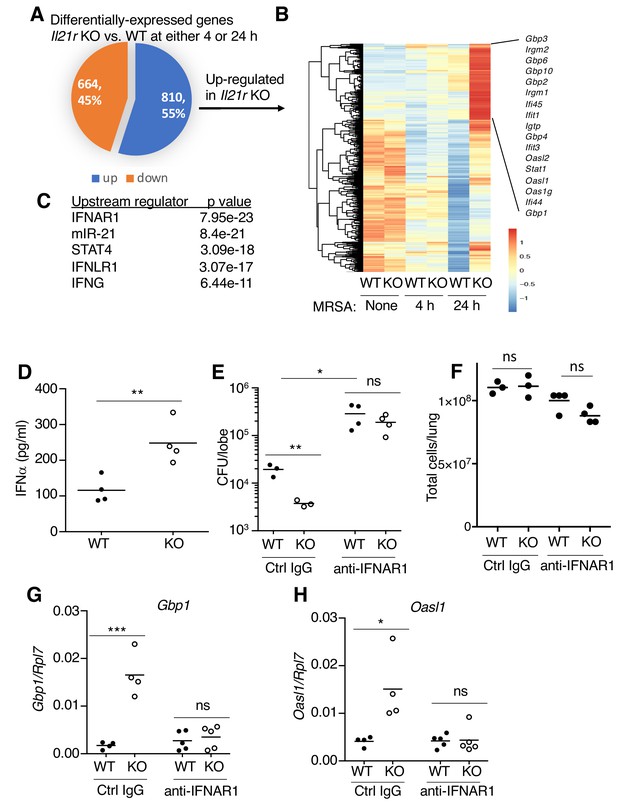
RNA-Seq analysis revealed an enhanced IFN profile in Il21r KO lungs.
(A) Venn diagram showing the number of differentially expressed genes (1474) in WT vs Il21r KO lungs, with 664 genes downregulated and 810 genes upregulated in the Il21r KO. (B) Heat map showing a cluster of IFN-related genes more highly expressed in Il21r KO than in WT lung. (C) Pathway analysis of differentially expressed genes shows enrichment for IFN-related genes. (D) IFNα was measured by ELISA in BAL fluid from WT and Il21r KO mice 4 hr after infection with MRSA. (E) Diminished MRSA killing in lungs at 24 hr in mice pre-treated with anti-IFNAR1. (F) Total lung cellularity at 24 hr in mice pre-treated with Ctrl IgG or anti-IFNAR; the differences observed were not statistically significant at p<0.05. (G, H) Blocking type I IFN signaling with anti-IFNAR1 prior to infection with MRSA prevented enhanced expression of Gbp1 (G) and Oasl1 (H) mRNAs at 24 hr in Il21r KO lungs. Expression was normalized to Rpl7, Representative results from one of 2 independent RNA-Seq experiments are shown in panel A-C; in D-H, each experiment was performed three times with similar results.
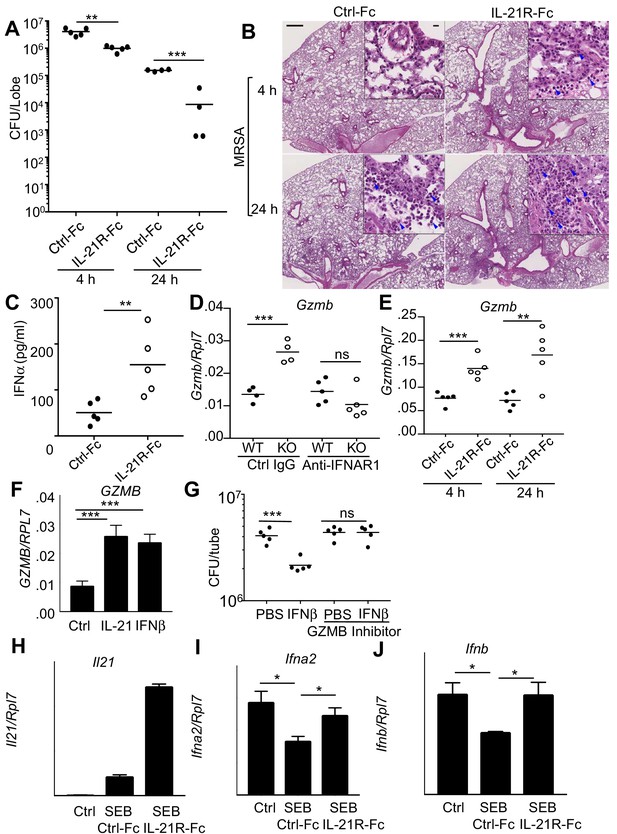
Blocking IL-21 signaling with IL-21R-Fc in WT mice phenocopies the enhanced MRSA killing seen in Il21r KO mice.
(A) WT mice were treated intratracheally with 50 μg of control Fc (from IgG1) or IL-21R-Fc protein for 2 days prior to infection with MRSA and lung CFU were quantitated at 4 and 24 hr post-infection with MRSA. (B) Histology of lungs in control Fc or IL-21R-Fc pre-treated mice at 4 and 24 hr post-infection (bar in upper left panel = 500 μm; bar in inset = 10 μm). (C) IFNα levels in BAL fluid were measured by ELISA 4 hr after infection in mice pretreated with control Fc or IL-21R-Fc. (D) Gzmb mRNA was measured in lungs of WT or Il21r KO mice pre-treated with isotype control or anti-IFNAR1 antibodies prior to MRSA infection, and normalized to Rpl7 mRNA expression. (E) Gzmb mRNA was measured in lungs of mice treated with either control Fc or IL-21R-Fc prior to MRSA infection and normalized to Rpl7 expression. (F) Like IL-21, IFNβ also induced GZMB mRNA in human peripheral blood neutrophils and normalized to RPL7 expression. (G) IFNβ induces increased in vitro killing of MRSA by human peripheral blood neutrophils and this was prevented by a granzyme B inhibitor, Z-AAD-CMK. (H–J) Co-cultures of CD4+ T cells and dendritic cells were stimulated with SEB either in the presence of control Fc or IL-21R-Fc, and mRNA was quantitated by RT-PCR after 24 hr and normalized to Rpl7 expression. Representative experiments are shown; each experiment was performed three times with similar results.
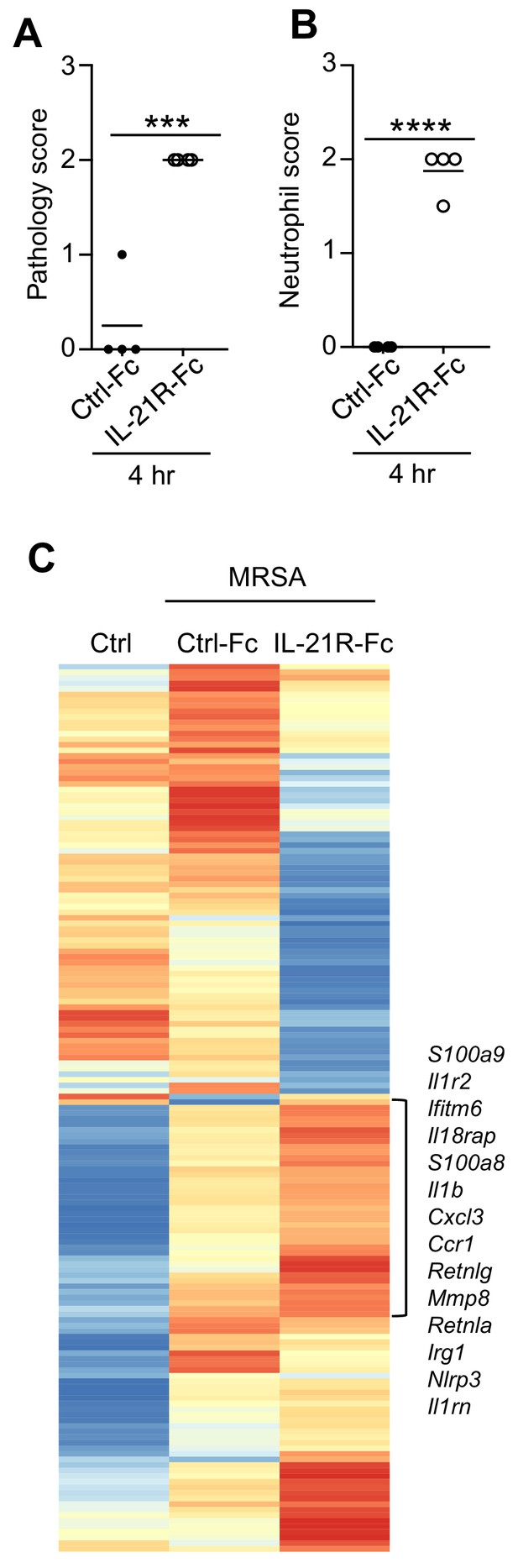
Pathology and RNA-Seq analysis of lungs from mice treated with control Fc or IL-21R-Fc prior to i.t.
MRSA infection, as detailed in the legend to Figure 6A. (A, B) Pathology and neutrophil infiltration scores were assessed from H and E-stained lung sections shown in Figure 6B. (C) RNA-Seq was performed on total lung mRNA either before infection (Ctrl) or at 4 hr after MRSA infection of mice pretreated with either Fc protein or an IL-21R-Fc fusion protein. Representative of 2 independent RNA-Seq experiments.
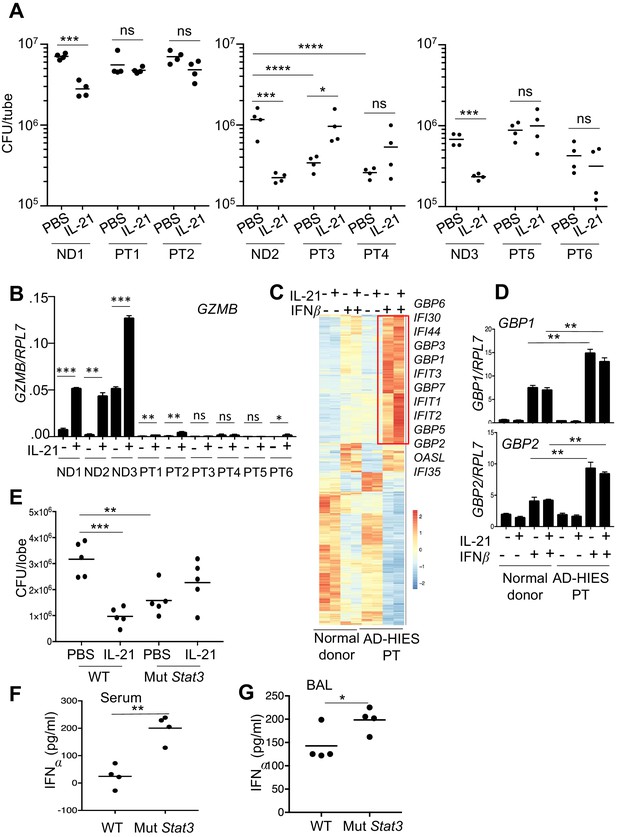
Neutrophils from patients with autosomal dominant hyper-IgE syndrome (AD-HIES) display reduced ex vivo IL-21-induced MRSA cytotoxic function.
(A) Peripheral blood neutrophils from normal donors (NDs) or AD-HIES patients (PTs) were assayed using an in vitro MRSA killing assay in the absence or presence of IL-21 (100 ng/ml). (B) RT-PCR was used to quantitate GZMB mRNA in neutrophils 4 hr after in vitro stimulation without or with IL-21. Expression was normalized to RPL7 expression (C) RNA-Seq was performed on normal donor and AD-HIES patient neutrophils stimulated for 4 hr with PBS, IL-21, IFNβ, or IL-21 +IFNβ. Genes differentially expressed (fold-change >1.5) in two independent RNA-Seq analyses are shown. (D) RT-PCR normalized to RPL7 expression was used to validate the expression pattern of GBP1 and GBP2 in neutrophils from additional normal donors and AD-HIES patients. (E–G) Mutant Stat3 transgenic mice were treated i.t. with PBS or 2 μg IL-21, infected i.t. 24 hr later with MRSA, and at 7 hr post-infection lung MRSA CFU quantitated (E), and IFNα levels were measured in the serum (F) and BAL fluid (G).
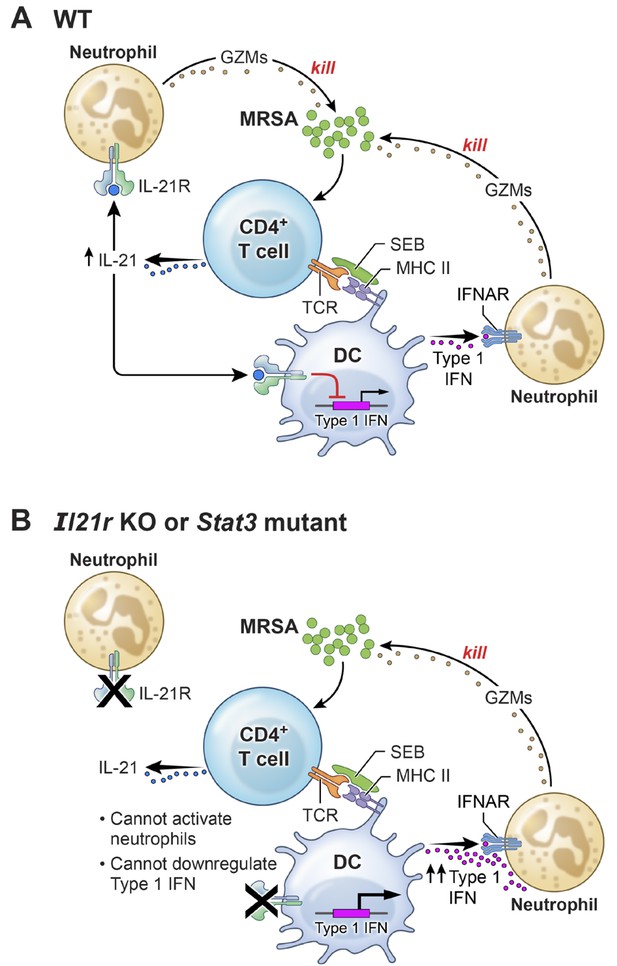
A model for the functional interplay of IL-21 with type I interferons in the response to MRSA.
(A) In wild-type (WT) mice, MRSA produces SEB, which bridges between T cells and dendritic cells by interacting with TCR and MHCII, leading to the production of IL-21 by CD4+ T cells. IL-21 stimulates neutrophils to release granzymes, with enhanced killing of MRSA. Type I IFN (e.g., produced by dendritic cells, as shown in the cartoon) can also induce granzyme production to promote killing of MRSA, but IL-21 inhibits dendritic cell production of type I IFN. (B) In the absence of either IL-21R or a functional STAT3 signaling response, the IL-21-induced neutrophil granzyme production does not occur. However, IL-21-mediated repression of type I IFN production by dendritic cells also no longer occurs, which leads to enhanced production of type I IFN and hence increased killing of MRSA through this pathway. The functional cross-talk of and relative potency of the IL-21- and type 1 IFN-mediated pathways and levels of each cytokine influence the outcome.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (S. aureus) | FPR3757 | ATCC | ATCC BAA-1556 | |
| Strain, strain background (Streptococcus pyogenes) | NZ131 | ATCC | ATCC-BAA-1633 | |
| Genetic reagent (M. musculus) | Il21r-/- | Ozaki et al. Science 298:1630, 2002 | ||
| Genetic reagent (M. musculus) | mCherry Il21 | Wang et al. P.N.A.S. 108:9542, 2011 | ||
| Genetic reagent (M. musculus) | C57BL/6-Tg(Stat3*)9199Alau/J | Jackson Laboratory | Stock # 027952 | |
| Biological sample (S. aureus) | Staph enterotoxin B | List Biological Laboratories | #122 | |
| Biological sample (S. aureus) | Heat-killed S. aureus | In Vivo Gen | tlrl-hksa | |
| Antibody | Mouse monoclonal anti-granzyme B | Biolegend | GB11 RRID:AB_2562195 | flow cytometry 1:100 |
| Antibody | Mouse monoclonal anti-granzyme B | Biolegend | clone M3304B06 RRID:AB_2565266 | WB (1:500) |
| Antibody | Mouse monoclonal anti-Ly6G (1A8) | BioX Cell | RRID: AB_1107721 | Blocking Ab |
| Antibody | Mouse monoclonal anti-IFNAR (MAR1-5A3) | BioX Cell | RRID: AB_2687723 | Blocking Ab |
| Antibody | Mouse monoclonal MOPC-21 | BioX Cell | RRID: AB_1107784 | Blocking Ab |
| Antibody | Rat monoclonal 2A3 | BioX Cell | RRID: AB_1107769 | Blocking Ab |
| Antibody | Mouse monoclonal anti-human IL21 | BD Biosciences | clone 3A3-N2.1 RRID:AB_2738933 | flow cytometry 1:100 |
| Antibody | Rat monoclonal anti-mouse IL-21R | BD Biosciences | clone 4A9 | flow cytometry 1:100 |
| Antibody | Mouse monoclonal anti-human IL-21R | Biolegend | clone 2G1-K12 RRID:AB_2123988 | flow cytometry 1:100 |
| Antibody | Mouse monoclonal anti-human CD66b | Biolegend | clone G10F5 RRID:AB_2563294 | flow cytometry 1:100 |
| Antibody | Mouse monoclonal anti-CD11b | Biolegend | clone M1/70 RRID:AB_312789 | flow cytometry 1:100 |
| Peptide, recombinant protein | Recombinant mouse IL-21 | R and D Systems | 594 ML-100/CF | |
| Peptide, recombinant protein | Recombinant human IL-21 | R and D Systems | 8879-IL-050/CF | |
| Peptide, recombinant protein | Recombinant human GM-CSF | R and D Systems | 215 GM | |
| Peptide, recombinant protein | Recombinant human IL-4 | R and D Systems | 204-IL | |
| Peptide, recombinant protein | Z-AAD-CMK | Enzo Life Sciences | BML-P165 | |
| Commercial assay or kit | IL-21 ELISA | R and D Systems | DY594 | |
| Commercial assay or kit | Granzyme B ELISA | Ebioscience | BMS2027 RRID:AB_2575322 | |
| Commercial assay or kit | IFNalpha ELISA | Ebioscience | 50-246-672 | |
| Commercial assay or kit | Cell Rox Deep Red staining kit | InVitrogen/Molecular Probes | C10491 | |
| Commercial assay or kit | Mouse neutrophil isolation kit | Miltenyi | 130-097-658 | |
| Commercial assay or kit | Human neutrophil isolation kit | Stem Cell Technologies | 19666 | |
| Commercial assay or kit | KAPA stranded mRNA-Seq library kit | Kapa Biosystems | KK8580 | |
| Chemical compound, drug | Peptidoglycan from S. aureus | Sigma | #77140 |
Additional files
-
Supplementary file 1
RNA-Seq analysis was performed on lung mRNA isolated 7 or 24 hr after treatment of WT mice with PBS or IL-21.
- https://doi.org/10.7554/eLife.45501.016
-
Supplementary file 2
RNA-Seq was performed on highly purified human neutrophils incubated for 4 or 24 hr with PBS or IL-21 in the absence or presence of heat-killed S. aureus.
- https://doi.org/10.7554/eLife.45501.017
-
Supplementary file 3
RNA-Seq analysis from WT and Il21r KO mouse lungs 4 and 24 hr after MRSA infection.
- https://doi.org/10.7554/eLife.45501.018
-
Supplementary file 4
RNA-Seq was performed on total lung mRNA either before MRSA infection or 4 hr after infection of mice that were pre-treated with either control Fc protein or IL-21R-Fc fusion protein.
- https://doi.org/10.7554/eLife.45501.019
-
Supplementary file 5
RNA-Seq analysis of neutrophils from normal human donors and AD-HIES patients; cells were either not treated or treated for 4 hr with IL-21, IFNβ, or IL-21 +IFNβ.
- https://doi.org/10.7554/eLife.45501.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.45501.021





