Caspase-mediated cleavage of IRE1 controls apoptotic cell commitment during endoplasmic reticulum stress
Figures
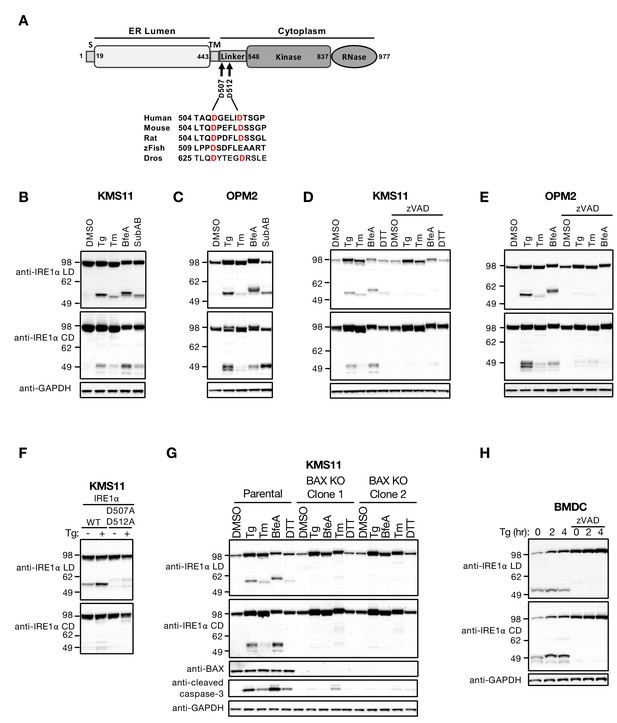
ER stress induces caspase-mediated cleavage of IRE1 in the cytoplasmic linker region.
(A) Schematic representation of the human IRE1 protein and comparison of the amino acid sequences surrounding the predicted caspase cleavage sites in the linker region of IRE1 from different species (zFish, zebrafish; Dros, Drosophila melanogaster). (B, C) KMS11 (B) or OPM2 (C) cells were treated with 100 nM Tg, 5 μg/ml Tm, or 0.2 μg/ml BfeA for 16 hr, or 0.3 μg/ml SubAB for 3 hr. Cell lysates were analyzed by western blot (WB) with anti-IRE1α LD or anti-IRE1α CD antibody to detect the lumenal or cytoplasmic domains. (D, E) KMS11 (D) or OPM2 (E) cells were treated with 100 nM Tg, 5 μg/ml Tm, 0.2 μg/ml BfeA, or 1 mM DTT for 16 hr in the absence or presence of 20 μM zVAD. Samples were analyzed as in B for the presence of cleavage products. (F) A cDNA plasmid expressing either WT or doubly mutated IRE1 (D507A, D512A) was transiently transfected into KMS11 cells harboring CRISPR/Cas9-based IRE1 knockout. Cells were treated with either DMSO or 100 nM Tg for 16 hr and analyzed by WB as indicated. (G) Two independent KMS11 clones harboring CRISPR/Cas9-based BAX knockout were generated and validated for BAX deletion as compared to the parental cell line. Cells were treated with DMSO, 100 nM Tg, 0.5 μg/ml BfeA, 5 μg/ml Tm, or 1 mM DTT for 24 hr and analyzed by WB. (H) BMDCs were obtained from C57/BL6 mice and treated with 100 nM Tg in the absence or presence of 20 μM zVAD for the indicated times. Equal amounts of protein from cell lysates were analyzed by WB. B–H show representative results from at least three similar experiments. DMSO vehicle was used as control.
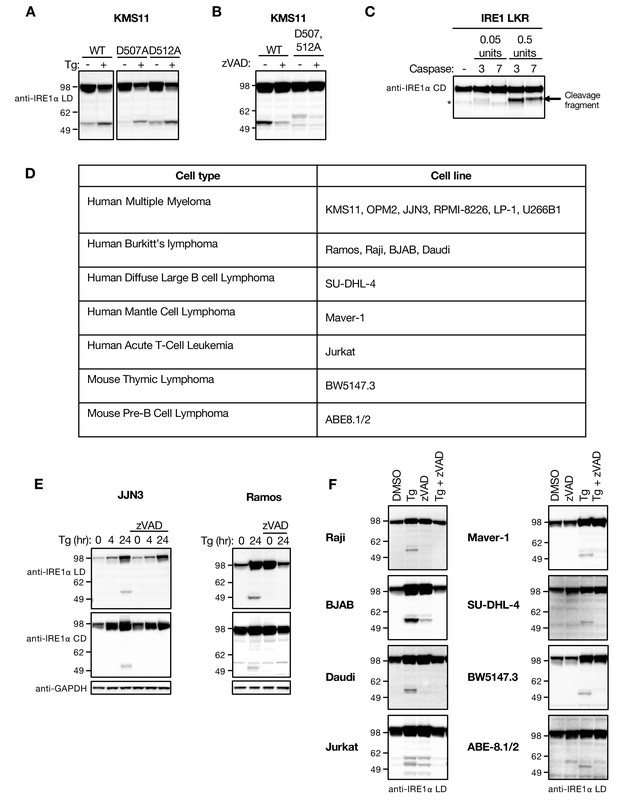
ER stress induces caspase-mediated cleavage of IRE1 in the cytoplasmic linker region.
(A) KMS11 cells harboring IRE1 knockout were transiently transfected with WT IRE1 or mutant IRE1 in which the Asp at position 507 or 512 was replaced by Ala. After 48 hr, cells were treated with DMSO or 100 nM Tg for another 24 hr and analyzed by WB. (B) KMS11 cells were transfected with a plasmid expressing either WT IRE1 or a D507A, D512A double mutant and analyzed as in A, in the absence or presence of 20 μM zVAD. (C) Purified caspase-3 or caspase-7 proteins were incubated with a purified recombinant IRE1 protein comprising the linker, kinase and RNase domains (IRE1 LKR) and analyzed by western blot. Asterisk indicates a background band, while arrow indicates the cleavage fragment. (D) Summary of cell lines that displayed similar IRE1 processing in response to treatment with Tg. (E, F) Corresponding WB data for most of the cell lines listed in the table. Cells were incubated with 100 nM Tg in the absence or presence of 20 μM zVAD for the indicated time (E) or 24 hr (F). WB data show representative results from at least three similar experiments. DMSO vehicle was used as control.
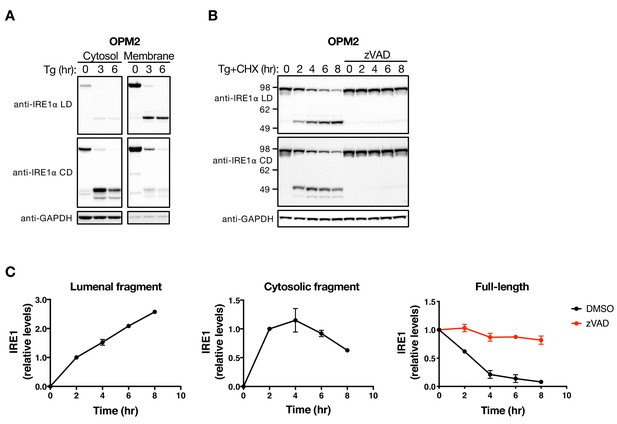
The LD and CD fragments of IRE1 differ in their cellular disposition.
(A) OPM2 cells were treated with 100 nM Tg for the indicated time, subjected to subcellular fractionation, and the cytosol and membrane fractions were analyzed by WB. (B) OPM2 cells were treated with 100 nM Tg to induce ER stress as well as 10 μg/ml CHX to block protein synthesis, in the absence or presence of 20 μM zVAD to block caspase activity. After the indicated incubation time, cells were lysed and analyzed by WB. (C) Levels of the lumenal or cytoplasmic fragments or full-length IRE1 (anti-IRE1α LD) relative to GAPDH were quantitated using ImageJ and plotted as indicated. Data represent mean with standard deviation (SD) from two independent experiments. DMSO vehicle was used as control.
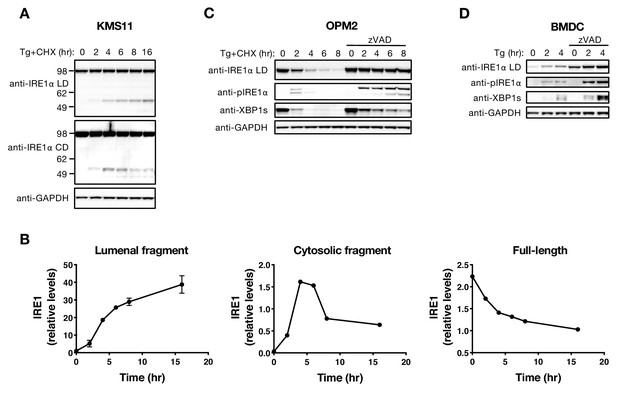
The LD and CD fragments of IRE1 differ in their cellular disposition.
(A) KMS11 cells were treated with 100 nM Tg and 10 μg/ml CHX for the indicated time and cells were harvested for protein isolation. Equal amounts of protein were analyzed by WB with the indicated antibodies. (B) Levels of the lumenal or cytoplasmic fragments or full-length IRE1 (anti-IRE1α LD) relative to GAPDH in panel A were quantitated using ImageJ and plotted as indicated. Data represent mean ± SD from two independent experiments. (C) OPM2 cells were treated with 100 nM Tg and 10 μg/ml CHX with or without 20 μM zVAD for the indicated time and cell lysates were analyzed by WB. (D) BMDC were treated with 100 nM Tg with or without 20 μM zVAD and cell lysates were analyzed by WB. WB data represent two or three independent experiments. DMSO vehicle was used as control.
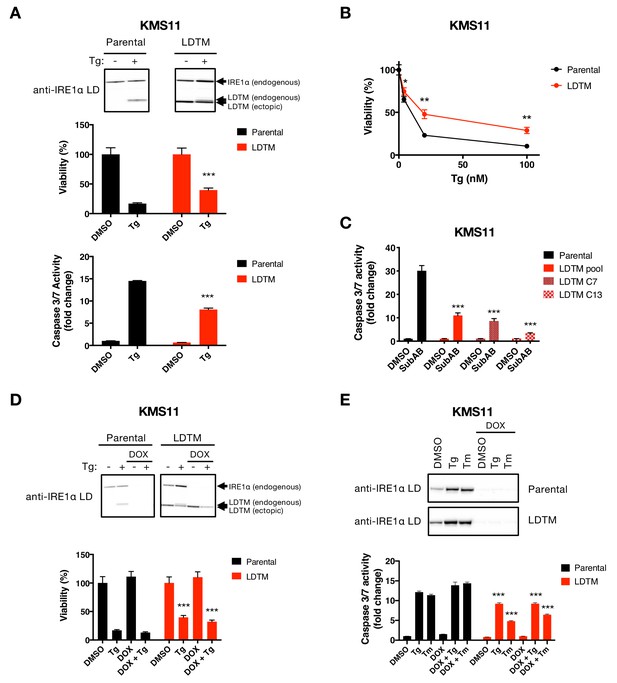
Ectopic expression of IRE1 LDTM attenuates apoptotic caspase activation independent of full-length IRE1.
(A) KMS11 parental cells or cells stably expressing a cDNA plasmid encoding LDTM (1-470) driven by the CMV promoter were treated with DMSO or 100 nM Tg for 24 hr. Cell viability was measured using CellTiter-Glo normalized by the number of cells at seeding (middle panel). The percentage of viable cells is graphed as an average of three biological replicates. Equal amounts of protein from cell lysates were analyzed by WB (top panel) or Caspase-Glo 3/7 assay (bottom panel). WBs are representative of two or more experiments and the graph depicts mean ± SD of three technical replicates. (B) KMS11 cells as in A were treated with different concentrations of Tg for 24 hr and analyzed for viability. The percentage of viable cells is graphed as an average of three biological replicates ± SD. (C) KMS11 parental cells, LDTM expressing KMS11, or single cell clones derived from the LDTM transfected KMS11 pool (C7 or C13) were treated with 0.3 μg/ml SubAB for 3 hr. Equal amounts of protein from cell lysates were analyzed using the Caspase-Glo 3/7 assay. The graph depicts mean luminescence signal normalized to the control ± SD of three technical replicates. (D) KMS11 cells were stably transfected with a DOX-inducible shRNA plasmid targeting IRE1 (Parental). The cells were then stably transfected with a cDNA plasmid encoding LDTM (1-470) driven by the CMV promoter as in A. Parental and ectopic LDTM-expressing cells were treated for 3 days in the absence or presence of 1 μg/ml DOX to induce shRNA-mediated depletion of endogenous IRE1. Cells were then treated with DMSO or 100 nM Tg for 24 hr to induce ER stress and analyzed by WB (top panel) or CellTiterGlo assay (bottom panel). The percentage of viable cells is graphed as an average of three biological replicates ± SD. (E) KMS11 cells expressing DOX-inducible IRE1 shRNA were treated in the absence or presence of 1 μg/ml DOX and then subjected to ER-stress induction with 100 nM Tg or 5 μg/ml Tm for 24 hr. Cells were analyzed by WB (top panel) or Caspase-Glo 3/7 assay (bottom panel). The graph depicts mean luminescence signal normalized to DMSO ± SD of three technical replicates.

Ectopic expression of IRE1 LDTM attenuates apoptotic caspase activation independent of full-length IRE1.
(A) KMS11 overexpressing an untagged version of LDTM (amino acids 1–470) or parental cells were treated with DMSO or 100 nM Tg for 24 hr. Equal amounts of protein from cell lysates were analyzed by Caspase-Glo 3/7 assay. Graphs depict mean luminescence signal ± SD as a ratio to the DMSO control from three technical replicates. (B) Two independent clones expressing ectopic LDTM (1-470) were isolated (C7 and C13). Cells were treated with different concentrations of Tg for 72 hr and viability was measured by CellTiter-Glo assay. The percentage of viable cells is graphed as an average of three biological replicates ± SD. (C) Parental KMS11 cells or cells stably transfected with a plasmid encoding a CMV-driven LDTM protein, representing a precise cleavage fragment of IRE1 with a Flag tag (1–507F), were treated with DMSO or 0.3 μg/ml SubAB for 3 hr and analyzed by Caspase-Glo 3/7 assay. The graph depicts mean luminescence signal ± SD as a ratio to the DMSO control from three technical replicates. (D) Parental JJN3 cells or JJN3 cells transfected as in C were treated with 100 nM Tg, or 5 μg/ml Tm, or 0.5 μg/ml BfeA for 24 hr, or 0.3 μg/ml SubAB for 3 hr, and analyzed by Caspase-Glo 3/7 assay. The graph depicts mean luminescence signal ± SD as a ratio to the DMSO control from three technical replicates. (E) Fixed MDA-MB-231 cells expressing the flag-tagged LDTM construct were stained with anti-flag (red), anti-calnexin (green), and DAPI. Cells were imaged using a confocal microscope and a z-stack image was acquired to determine localization of LDTM in the ER. An orthogonal projection is shown. (F, G) KMS11 cells expressing DOX-inducible IRE1 shRNA (Parental) were stably transfected with a plasmid encoding a CMV-driven LDTM protein without (1-507) or with a Flag tag (1–507F). The cells were incubated in the absence or presence of 1 μg/ml DOX for 3 days to induce IRE1 shRNA expression. Cells were then treated with DMSO, 100 nM Tg, or 5 μg/ml Tm for 6 hr and equal amounts of protein lysates were analyzed by Caspase-Glo 3/7 assay (F) or WB (G). Data represent at least three independent experiments; graphs depict mean ± SD of three technical replicates.
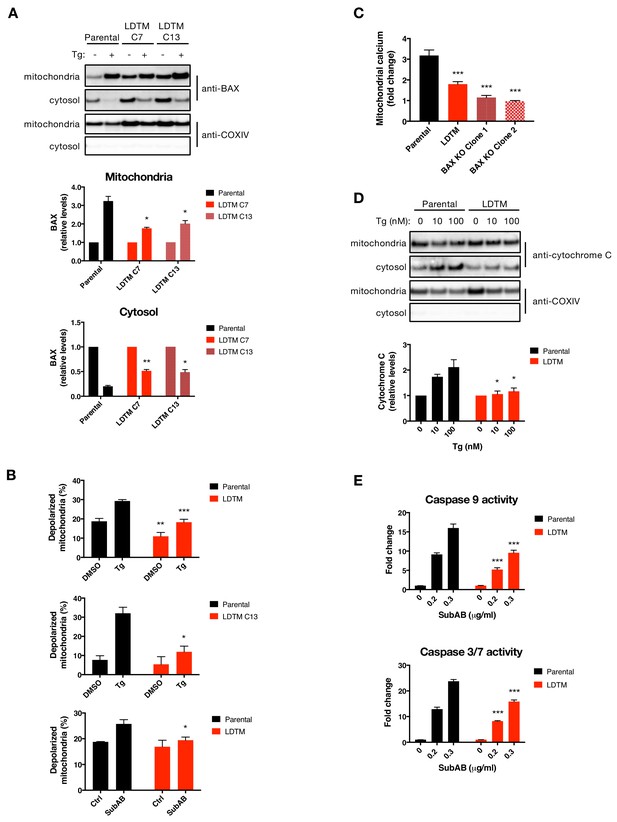
LDTM attenuates key mitochondrial apoptotic events.
(A) Parental KMS11 cells or two clones expressing ectopic LDTM (1-470) were treated with DMSO or 100 nM Tg for 20 hr. Cells were differentially lysed to enrich for mitochondrial or cytoplasmic fractions and equal amounts of protein were analyzed by WB (top). Mitochondrial BAX levels were quantitated by ImageJ relative to the mitochondrial marker COXIV; cytosolic levels were similarly quantitated and graphed in relation to the corresponding DMSO controls. Data represent mean ± SD from two independent experiments. (B) Parental KMS11 cells or LDTM overexpressing cells, either a pool (top panel) or clone 13 (middle panel) were treated with 100 nM Tg for 20 hr. Similarly, parental cells and the LDTM overexpressing pool were treated with 0.3 μg/ml SubAB for 3 hr (bottom). Cells were subsequently incubated with 2 μM JC-1 dye for 30 min and analyzed for mitochondrial depolarization by FACS based on a fluorescence emission shift from red (~590 nm) to green (~529 nm). The average percentage of cells exhibiting mitochondrial depolarization ± SD from two or more biological replicates is graphed. (C) Parental KMS11 cells, LDTM overexpressing cells or two cell lines harboring a BAX deletion were treated with 100 nM Tg for 24 hr. Cells were incubated with the mitochondrial calcium dye Rhod-2 and then analyzed by FACS. Data represent the mean fold change in fluorescence ± SD as compared to DMSO treated cells from three or more biological replicates. (D) Parental KMS11 cells or cells expressing ectopic LDTM (1-470) were treated with DMSO or 10 or 100 nM Tg for 20 hr and differentially lysed to enrich for mitochondrial or cytoplasmic protein. Equal amounts of protein were analyzed by WB (top) and cytosolic amounts of cytochrome C were quantitated by ImageJ and graphed relative to the corresponding DMSO controls (bottom). Bar graphs represent mean ± SD from two independent experiments. (E) Parental KMS11 cells or cells expressing ectopic LDTM (1-470) were treated with 0.2 or 0.3 μg/ml SubAB for 3 hr and analyzed by Caspase-Glo 9 (top) or Caspase-Glo 3/7 (bottom) assay. Graphs depict mean ± SD of three technical replicates.
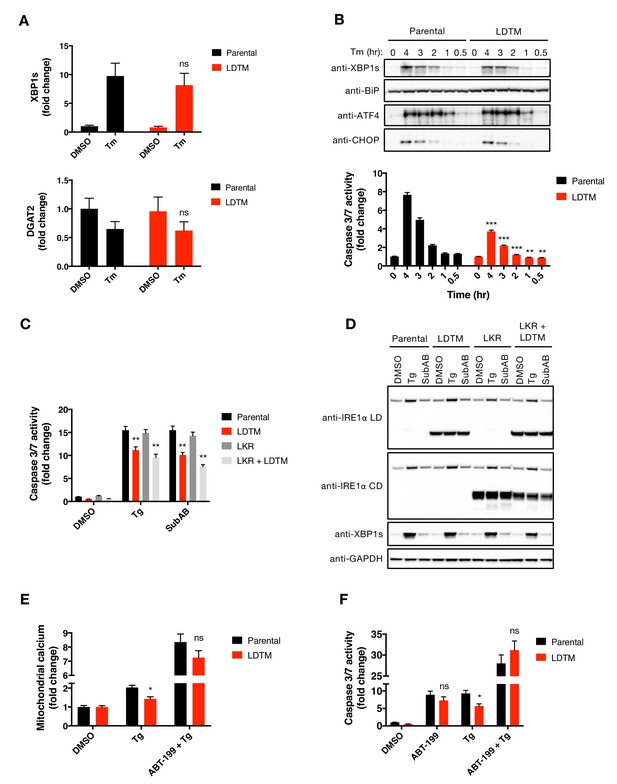
LDTM attenuates caspase activation without affecting the UPR and its function is reversed by BCL2 inhibition.
(A) Parental KMS11 cells or cells expressing ectopic LDTM (1-470) were treated with DMSO or 5 μg/ml Tm for 6 hr. RNA isolated from the cells was analyzed by RT-qPCR for the expression of XBP1s (top) or DGAT2 (bottom) transcripts. Graphs depict mean ± SD of three technical replicates. (B) Cells as in A were treated with DMSO or 5 μg/ml Tm for the indicated amount of time and analyzed by WB (top) or Caspase-Glo 3/7 assay (bottom). WBs are representative of two or more experiments and the graph depicts mean ± SD of three technical replicates. (C, D) KMS11 cells expressing LDTM (1-470), LKR (468-977), or LDTM and LKR together were treated with either 100 nM Tg for 6 hr or 0.3 μg/ml SubAB for 3 hr. Equal amounts of protein from cell lysates were analyzed by Caspase-Glo 3/7 assay (C) or by WB (D). WBs are representative of two or more independent experiments and the graph depicts mean ± SD of three technical replicates. (E) Parental KMS11 cells or LDTM (1-470) overexpressing cells were treated with DMSO, 100 nM Tg, or 1 μM ABT-199 and 100 nM Tg for 20 hr. Cells were incubated with the mitochondrial calcium dye Rhod-2 and then analyzed by FACS. Data represents the fold change in fluorescence from the DMSO control from three or more biological replicates. (F) Parental KMS11 cells or LDTM (1-470) overexpressing cells were treated with DMSO, 1 μM ABT-199, 100 nM Tg, or 1 μM ABT-199 and 100 nM Tg for 20 hr. Equal amounts of protein from cell lysates were analyzed with Caspase-Glo 3/7 assay. The graph depicts mean luminescence as a ratio to the DMSO control ± SD of three technical replicates.
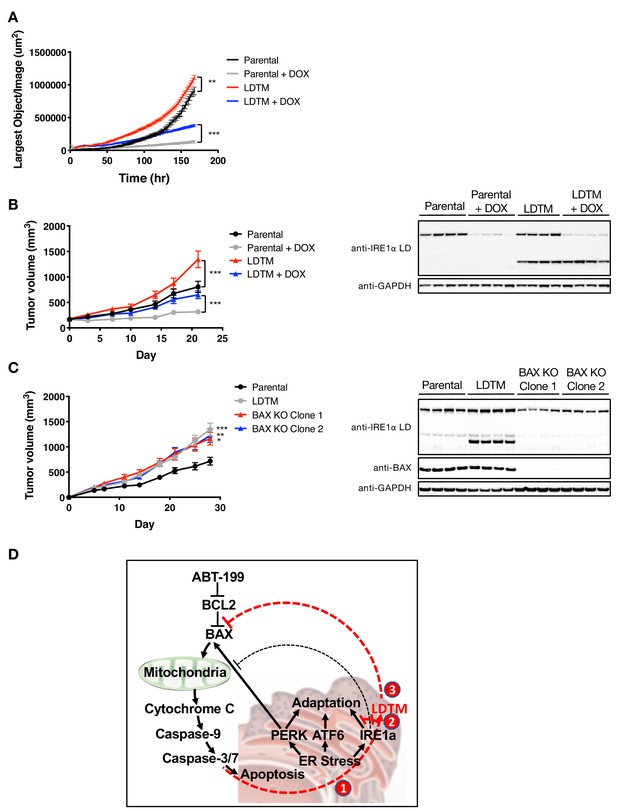
LDTM augments MM tumor progression.
(A) Parental KMS11 cells or cells expressing ectopic LDTM (1-470) were plated on matrigel and growth was monitored by the changes in confluence using an IncuCyte S3 instrument over 7 days. (B) KMS11 parental cells expressing DOX-inducible IRE1 shRNA or the same cells transfected with a plasmid encoding CMV-driven LDTM (1-470) were injected subcutaneously into CB-17 SCID mice. When tumors reached ~150 mm3 in volume, mice were divided into groups (n = 9) and treated with sucrose or DOX via the drinking water and tumor growth was monitored over 21 days. Tumors were harvested and lysates were analyzed by WB. (C) KMS11 parental (n = 20), LDTM overexpressing cells (n = 10), or two independent KMS11 clones harboring CRISPR/Cas9-based BAX knockout (n = 10 each) were injected subcutaneously into CB-17 SCID mice. Tumor growth was monitored over 28 days. After which, tumors were harvested and lysates were analyzed by WB. (D) Schematic model illustrating previously known (black text and lines) and novel (red text and lines) cross-regulation between the UPR and the apoptotic cascade. ER-stress-induced apoptotic signaling leads to caspase-dependent cleavage of IRE1 (1). This separates the sensing and signaling domains of IRE1, which dampens IRE1’s known XBP1s- and RIDD-mediated cytoprotective activities (2). Furthermore, it generates a fragment containing the lumenal domain and transmembrane segment (LDTM), which in turn suppresses further apoptotic signaling by attenuating BAX translocation to mitochondria (3) in a manner that can be reversed by the BCL2 inhibitor ABT-199.
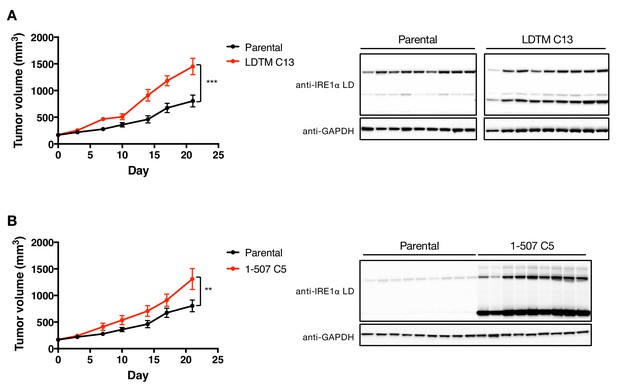
LDTM augments MM tumor progression.
(A) Parental KMS11 cells or clone 13 (C13) cells expressing ectopic LDTM (1-470) were injected subcutaneously into CB-17 SCID mice (Parental n = 9, LDTM C13 n = 10) and tumor growth was monitored over 21 days. Tumors were harvested and lysates were analyzed by WB. (B) Parental KMS11 cells or a clone of LDTM (1-507) expressing cells were injected subcutaneously into CB-17 SCID mice (Parental n = 9, 1–507 C5 n = 8) and tumor growth was monitored over 21 days. Tumors were harvested and lysates were analyzed by WB.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (mouse) | CB-17 SCID | Charles River Laboratories | ||
| Strain, strain background (mouse) | C57BL/6 | Charles River Laboratories | ||
| Cell line (human) | KMS11 | Genentech | RRID:CVCL_2989 | Multiple myeloma |
| Cell line (human) | OPM2 | Genentech | RRID:CVCL_1625 | Multiple myeloma |
| Cell line (human) | JJN3 | Genentech | RRID:CVCL_2078 | Multiple myeloma |
| Cell line (human) | RPMI-8226 | Genentech | RRID:CVCL_0014 | Multiple myeloma |
| Cell line (human) | LP-1 | Genentech | RRID:CVCL_0012 | Multiple myeloma |
| Cell line (human) | U266B1 | Genentech | RRID:CVCL_0566 | Multiple myeloma |
| Cell line (human) | Ramos | Genentech | RRID:CVCL_0597 | Burkitt’s lymphoma |
| Cell line (human) | Raji | Genentech | RRID:CVCL_0511 | Burkitt’s lymphoma |
| Cell line (human) | BJAB | Genentech | RRID:CVCL_5711 | Burkitt’s lymphoma |
| Cell line (human) | Daudi | Genentech | RRID:CVCL_0008 | Burkitt’s lymphoma |
| Cell line (human) | Maver-1 | Genentech | RRID:CVCL_1831 | Mantle Cell Lymphoma |
| Cell line (human) | Jurkat | Genentech | RRID:CVCL_0367 | Acute T-Cell Leukemia |
| Cell line (human) | MDA-MB-231 | Genentech | RRID:CVCL_0062 | Triple negative breast cancer |
| Cell line (mouse) | BW5147.3 | Genentech | RRID:CVCL_4135 | Thymic Lymphoma |
| Cell line (mouse) | ABE8.1/2 | Genentech | RRID:CVCL_3487 | Pre-B Cell Lymphoma |
| Cell line (human) | BAX knockout Clone one and Clone 2 | This paper | Isolated clones of CRISPR/Cas9 deletion of BAX gene in KMS11 | |
| Cell line (human) | IRE1 knockout KMS11 | Genentech | Harnoss et al., 2019 | |
| Biological sample (mouse) | BMDC | This paper | Isolated from the tibia and femur bones of C57BL/6 mice | |
| Transfected construct (human) | LDTM (1-470) | This paper | IRE1 aa1-470 with or without C-terminal Flag tag under CMV promoter | |
| Transfected construct (human) | 1-507/1-507F | This paper | IRE1 aa1-507 with or without C-terminal Flag tag under CMV promoter | |
| Transfected construct (human) | IRE1 shRNA | Genentech | Harnoss et al., 2019 | |
| Transfected construct (human) | IRE1 wt | This paper | Full-length wild-type IRE1 expressed from CMV promoter | |
| Transfected construct (human) | IRE1 D507A | This paper | Asp at 507 was mutated to Ala | |
| Transfected construct (human) | IRE1 D512A | This paper | Asp at 512 was mutated to Ala | |
| Transfected construct (human) | IRE1 D507A, D512A | This paper | Both Asp at 507 and 512 were mutated to Ala | |
| Transfected construct (human) | IRE1 LKR | This paper | IRE1 aa468-977 with N-terminal His tag expressed from CMV promoter | |
| Transfected construct (human) | BAX gRNA_1 | This paper | GCGGTGATGGACGGGTCCG | |
| Transfected construct (human) | BAX gRNA_2 | This paper | TTCATGATCTGCTCAGAGC | |
| Antibody | GAPDH-HRP | Cell Signaling Technology | Cat. #: 2118 | (1:5000) |
| Antibody | XBP1s (rabbit monoclonal) | Genentech (Chang et al., 2018) | (1:1000) | |
| Antibody | IRE1α LD (mouse monoclonal, IgG2a) | This study | (1:1000) | |
| Antibody | IRE1α CD (rabbit monoclonal) | Cell Signaling Technology | Cat. #: 3294 | (1:1000) |
| Antibody | BAX (rabbit polyclonal) | Cell Signaling Technology | Cat. #: 2772 | (1:1000) |
| Antibody | Cytochrome-C (rabbit monoclonal) | Cell Signaling Technology | Cat. #: 11940 | (1:1000) |
| Antibody | COXIV (rabbit monoclonal) | Cell Signaling Technology | Cat. #: 4850 | (1:1000) |
| Antibody | ATF4 (rabbit monoclonal) | Cell Signaling Technology | Cat. #: 11815 | (1:1000) |
| Antibody | BiP (rabbit monoclonal) | Cell Signaling Technology | Cat. #: 3177 | (1:1000) |
| Antibody | Cleaved caspase-3 (rabbit monoclonal) | Cell Signaling Technology | Cat. #: 9664 | (1:1000) |
| Antibody | CHOP (mouse monoclonal IgG2a) | Cell Signaling Technology | Cat. #: 2895 | (1:1000) |
| Antibody | Flag (mouse monoclonal IgG1) | Sigma-Aldrich | Cat. #: F1804 | (1:1000) |
| Antibody | Calnexin (rabbit polyclonal) | Abcam | Cat. #: ab22595 | (1 μg/ml) |
| Antibody | Anti-rabbit IgG HRP | Jackson ImmunoResearch Laboratories | Cat. #: 711-035-152 | (1:10,000) |
| Antibody | Anti-mouse IgG2a HRP | SouthernBiotech | Cat. #:1080–05 | (1:10,000) |
| Antibody | pIRE1α (rabbit monoclonal) | Genentech (Chang et al., 2018) | (1:500) | |
| Recombinant DNA reagent | pRK.TK.Neo-IRE1 WT | Genentech | IRE1 wild-type cDNA in pRK.TK.Neo backbone | |
| Recombinant DNA reagent | pRK.TK.Neo-IRE1 D507A | Genentech | IRE1 cDNA with Asp 507 mutated to Ala in pRK.TK.Neo backbone | |
| Recombinant DNA reagent | pRK.TK.Neo-IRE1 D512A | Genentech | IRE1 cDNA with Asp 512 mutated to Ala in pRK.TK.Neo backbone | |
| Recombinant DNA reagent | pRK.TK.Neo-IRE1 D507A, D512A | Genentech | IRE1 cDNA with Asp 507 and 512 mutated to Ala in pRK.TK.Neo backbone | |
| Recombinant DNA reagent | pRK.TK.Neo-IRE1 1–470-Flag | Genentech | IRE1 cDNA aa1-470 with C-terminal Flag in pRK.TK.Neo backbone | |
| Recombinant DNA reagent | pRK.TK.Neo-IRE1 1–470 | Genentech | IRE1 cDNA aa1-470 in pRK.TK.Neo backbone | |
| Recombinant DNA reagent | pRK.TK.Neo-IRE1 1–507-Flag | Genentech | IRE1 cDNA aa1-507 with C-terminal Flag in pRK.TK.Neo backbone | |
| Recombinant DNA reagent | pRK.TK.Neo-IRE1 1–507 | Genentech | IRE1 cDNA aa1-507 in pRK.TK.Neo backbone | |
| Recombinant DNA reagent | pcDNA3.1.Zeo-IRE1 6xHis 468–977 | Genentech | IRE1 cDNA aa468-977 with N-terminal His tag in pcDNA3.1.Zeo backbone | |
| Commercial assay or kit | Caspase-Glo 3/7 Assay | Promega | G8090 | |
| Commercial assay or kit | Caspase-Glo 9 Assay | Promega | G8210 | |
| Commercial assay or kit | CellTiter-Glo Luminescent Cell Viability Assay | Promega | G7570 | |
| Commercial assay or kit | MitoProbe JC-1 Assay Kit for Flow Cytometry | ThermoFisher Scientific | M34152 | |
| Commercial assay or kit | Subcellular Protein Fractionation Kit for Cultured Cells | ThermoFisher Scientific | 78840 | |
| Commercial assay or kit | Mitochondria Isolation Kit for Cultured Cells | ThermoFisher Scientific | 89874 | |
| Recombinant protein | LKR | This paper | Purified N-terminally His-tagged IRE1 aa468-977 | |
| Recombinant protein | Caspase 3 | Enzo Life Sciences | ALX-201–059 | |
| Recombinant protein | Caspase 7 | BioVision | 1087 | |
| Chemical compound, drug | Rhod-2, AM, cell permeant | Invitrogen | R1244 | |
| Chemical compound, drug | Thapsigargin, Tg | Tocris | 1138 | |
| Chemical compound, drug | Tunicamycin, Tm | Tocris | 3516 | |
| Chemical compound, drug | Brefeldin A, BfeA | Tocris | 1231 | |
| Chemical compound, drug | DTT | ThermoFisher Scientific | R0861 | |
| Chemical compound, drug | Cycloheximide, CHX | Sigma-Aldrich | C4859 | |
| Chemical compound, drug | Z-VAD-FMK, zVAD | R and D Systems | FMK001 | |
| Chemical compound, drug | Doxycycline, DOX | Clontech | NC0424034 | |
| Chemical compound, drug | ABT-199 | Genentech | G00376771 | |
| Chemical compound, drug | Subtilase toxin AB5, SubAB | Paton et al., 2006 | ||
| Software, algorithm | Prism 7 | GraphPad | ||
| Software, algorithm | ImageJ | NIH | ||
| Software, algorithm | FlowJo 10.4 | FlowJo, LLC |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47084.012





