Nucleolar dynamics and interactions with nucleoplasm in living cells
Figures
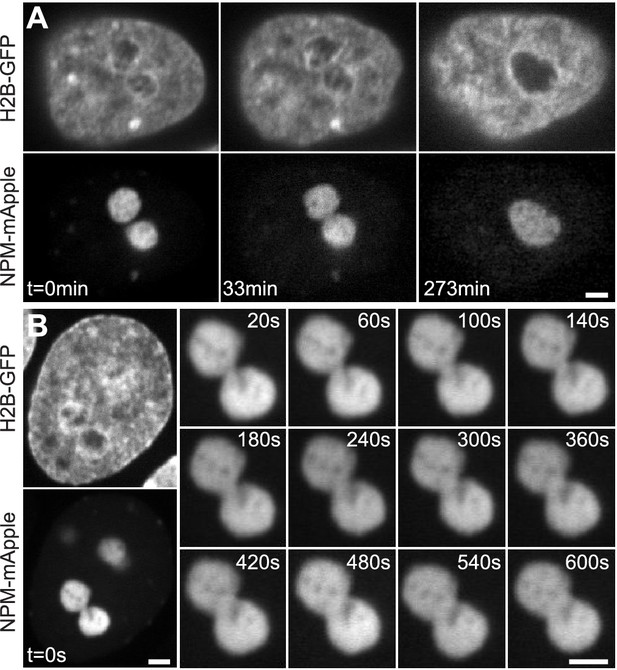
Nucleolar coalescence.
(A) Time lapse of a nucleus with fluorescently labeled chromatin (H2B-GFP) and two fusing nucleoli (NPM-mApple). Time points depict: pre-fusion ( = 0 min), with two distinct nucleoli, fusion in progress ( = 33 min), with a clearly visible neck connecting the two nucleoli, and post-fusion ( = 273 min), where a resultant nucleolus can be seen still rounding up. (B) Time series showing the growth of the neck connecting two coalescing nucleoli. At = 0 s, both the fluorescently labeled chromatin (H2B-GFP) and coalescing nucleoli (NPM-mApple) are depicted, the later frames, 20–600 s, show the progress of the nucleolar coalescence (NPM-mApple). Parts of (B) adapted from Figure 3b in Caragine et al. (2018). Scale bar, 2 µm.
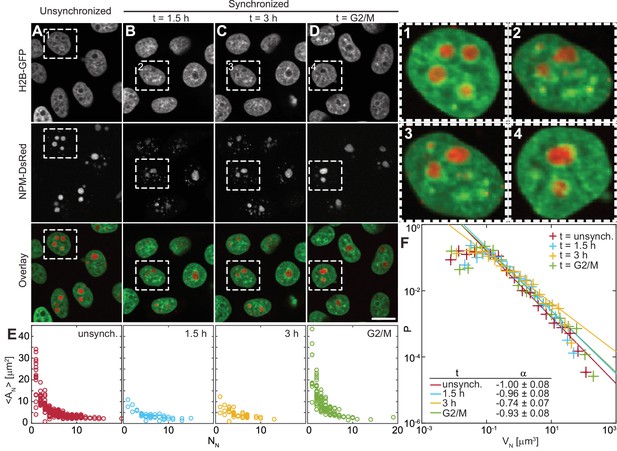
Nucleolar size distribution as a function of cell cycle.
(A–D) Micrographs of HeLa cell nuclei with fluorescently labeled chromatin (H2B-GFP) and nucleoli (NPM-DsRed) under the following conditions: unsynchronized cells (A), synchronized cells 1.5 hr (B) and 3 hr (C) after mitosis, and cells arrested at G2/M checkpoint (D). (1–4) enlarged view of the boxed nuclei from (A–D). (E) Average nucleolar area as a function of number of nucleoli per nucleus for all conditions from (A–D). For unsynchronized cells, total number of nucleoli analyzed = 1331 in 228 nuclei, for = 1.5 hr, = 275 in 42 nuclei, for = 3 hr, = 257 in 51 nuclei, and for = G2/M, = 497 in 124 nuclei. (F) Distributions of nucleolar volume and their fit to for all conditions from (A–D). For all fits, the goodness-of-fit, > 0.98. Scale bar, 15 µm.
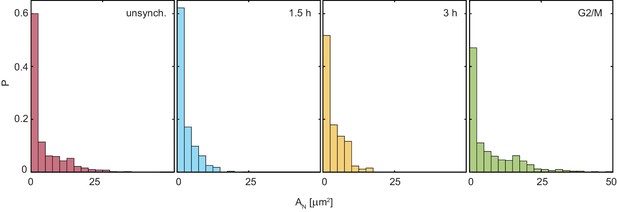
Nucleolar area distributions, , for all conditions shown in Figure 2E: unsynchronized cells, synchronized cells 1.5 hr after mitosis, synchronized cells 3 hr after mitosis, and cells arrested in G2/M.
For unsynchronized cells, number of analyzed nucleoli is = 1331 in 228 nuclei, for = 1.5 hr, = 275 in 42 nuclei, for = 3 hr, = 257 in 51 nuclei, and for = G2/M, = 497 in 124 nuclei.
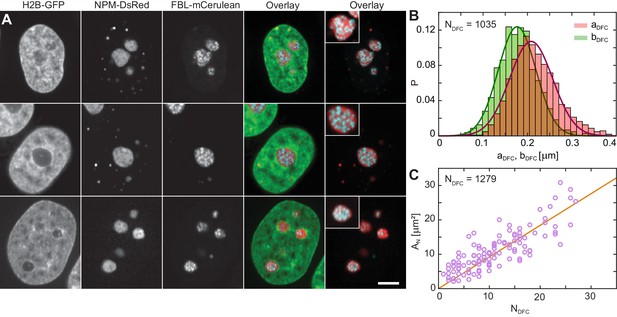
Nucleolar internal structure.
(A) Micrographs of HeLa nuclei with fluorescently labeled chromatin (H2B-GFP, green), nucleolar granular component (NPM-DsRed, red) and nucleolar dense fibrillar component (DFC) (FBL-mCerulean, blue) and overlays of all three signals (green, red, blue) and red and blue signal. The insets in overlay images present an enlarged view of a nucleolus from the image. (B) Distributions measured for semi-major axis (red) and semi-minor axis (green) of single DFCs ( = 1035). The solid red and green lines correspond to the Gaussian fits of distributions of and , respectively, with ≈ 210 nm and ≈ 180 nm. (C) Nucleolar area, , as a function of DFC number per nucleolus, , with a linear fit ≈ 0.92. We evaluated 1279 DFCs over 114 nucleoli in 63 nuclei. Scale bar, 5 µm.
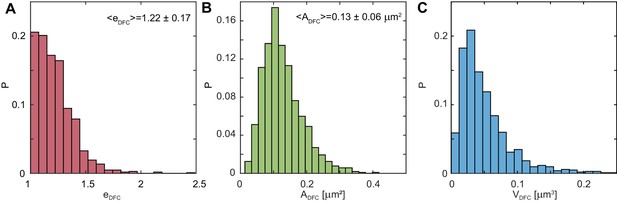
Distributions of DFC eccentricity, area and volume.
(A) Distribution of the measured eccentricities, , where and are the DFC semi-major and semi-minor axes, respectively ( 1035). (B) Distribution of the DFC area, , measured for single DFCs ( 1035). (C) DFC volume distribution, . DFC volume, , was calculated as , where is the semi-major axis of the DFC, providing the upper boundary on the volume estimate. exhibits a sharp maximum at = 0.030 μm3, suggesting largely a monodisperse population of DFCs ( 1035).
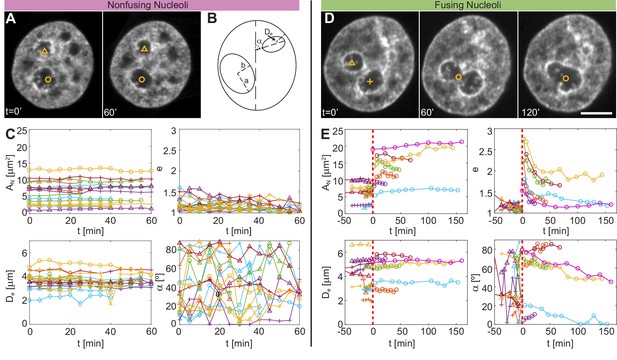
Comparison of size, shape and nuclear positioning between fusing and nonfusing nucleoli.
(A) Micrographs of a nucleus with fluorescently labeled chromatin (H2B-GFP), where two void spaces (labeled by yellow triangle and yellow circle) correspond to two nucleoli that did not fuse between = 0 and 60 min. (B) Schematics of measured variables. (C) Measured variables for nonfusing nucleoli: nucleolar area, , nucleolar eccentricity, , shortest distance from the nucleolar centroid to the nuclear envelope, , and the angle between the major nuclear and nucleolar axes, ( = 17, = 6). All characteristics are calculated in the nucleolar focal plane. (D) Micrographs of a nucleus with fluorescently labeled chromatin (H2B-GFP), where two void spaces (labeled by yellow triangle and yellow cross) correspond to two nucleoli before fusion at = 0 min, while at = 60 and 120 min they can be seen fusing (yellow circle). (E) Measured variables for fusing nucleoli: , , and ( = 12, = 7). The dashed red line at = 0 min indicates fusion. All measurements are carried out in the nucleolar focal plane. Scale bar, 5 µm.
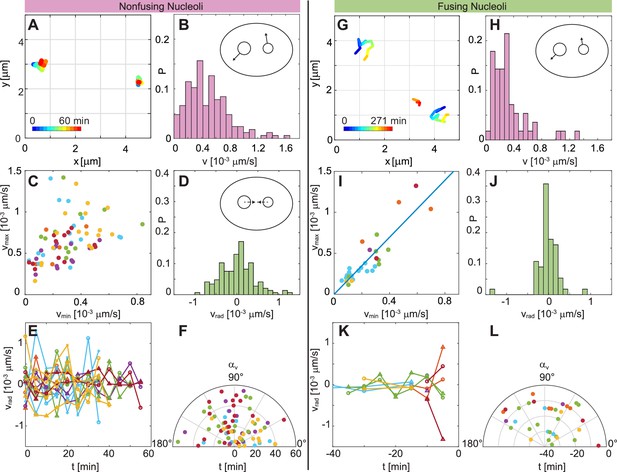
Comparison of dynamics between fusing and nonfusing nucleoli.
(A) Trajectories of two nonfusing nucleoli color-coded by their temporal evolution (blue to red). The time step is 5 min. (B) Histogram of the velocity magnitude, , for the nonfusing nucleoli ( = 17, = 6). (C) Velocities for pairs of nonfusing nucleoli, where and is the larger and the smaller nucleolar velocity, respectively. (D) Histogram of radial velocity, , for nonfusing nucleoli, with calculated with respect to the midpoint distance between nucleoli. (E) as a function of time for nonfusing nucleoli. (F) Velocity angle, , in polar coordinates as a function of time for nonfusing nucleoli. (G) Trajectories of pair of fusing nucleoli color-coded by their temporal evolution (blue to red). The pre-fusion nucleoli are visible at earlier times (blue to yellow), while the post-fusion nucleolus appears at later times (orange to red). The time step is 15 and 16 min. (H) Histogram of the for the fusing nucleoli ( = 12, = 7). (I) Velocities for pairs of fusing nucleoli, where and is the larger and the smaller nucleolar velocity, respectively, with a linear fit 1.74. (J) Histogram of for fusing nucleoli. (K) as a function of time for fusing nucleoli. (L) for fusing nucleoli as a function of time.
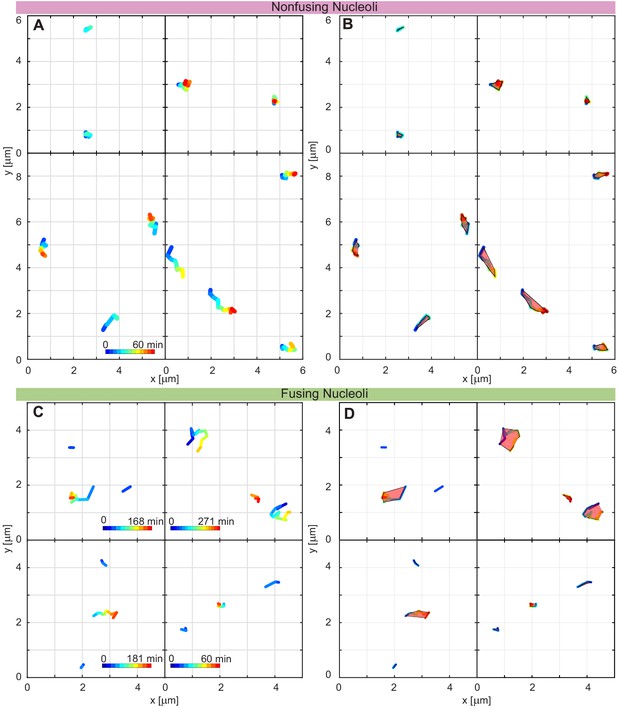
Additional nucleolar trajectories and enlarged view of nucleolar trajectories from Figure 5A and Figure 5G.
(A) Trajectories of nucleoli which do not fuse, time increases from blue to red (enlarged view of Figure 5A). (B) The trajectory areas determined as the convex hull of the trajectories of nucleoli from A. (C) Trajectories of nucleoli which fuse, time increases from blue to red (enlarged view of Figure 5G). The post fusion nucleolus is in between the other two trajectories. (D) The trajectory areas determined as the convex hull of the trajectories of nucleoli from C.
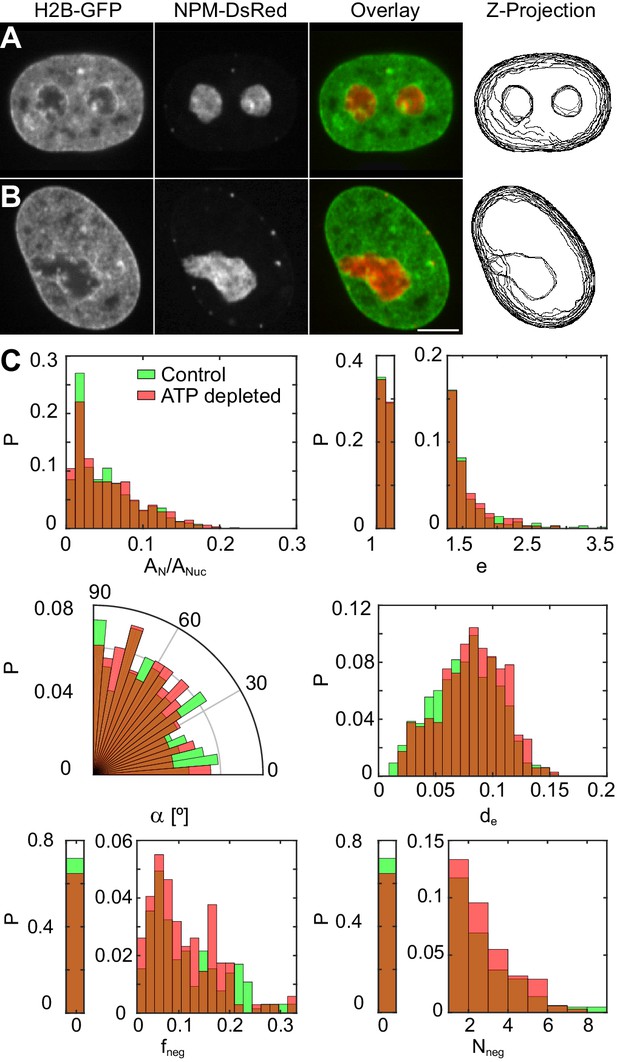
Nucleoli under physiological conditions and upon ATP-depletion.
(A–B) Micrographs of HeLa nuclei with fluorescently labeled chromatin (H2B-GFP, green) and nucleoli (NPM-DsRed, red), their color overlay and z-projections of nucleolar and nuclear contours: (A) under physiological conditions (control) and (B) after ATP depletion. (C) Distributions of the following nucleolar measurements under physiological conditions ( 648, 208) vs. upon ATP depletion ( 345, 127): nucleolar area normalized by nuclear area, , nucleolar eccentricity, , angle between the major nuclear and nucleolar axes, , the shortest distance from the nucleolar centroid to the nuclear envelope normalized by the nuclear circumference, , fraction of the nucleolar contour with negative curvature, , and number of continuous regions of negative curvature along the nucleolar contour, . All measurements are carried out in the nucleolar focal plane. Scale bar, 5 µm.
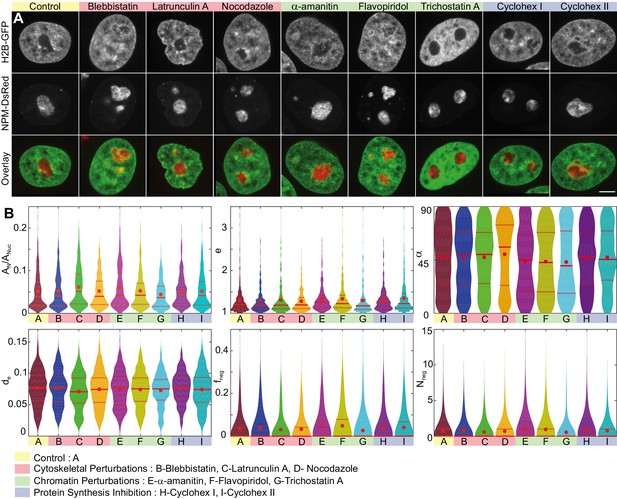
Nucleoli upon biochemical perturbations.
(A) Micrographs of HeLa nuclei with fluorescently labeled chromatin (H2B-GFP, green) and nucleoli (NPM-DsRed, red), and the overlay, under the following conditions: control, upon addition of blebbistatin, latrunculin A, nocodazole, -amanitin, flavopiridol, trichostatin A, and cycloheximide (at 30 min and 6.5 hr). (B) Histograms of the following measurements for all conditions (width indicates probability): nucleolar area normalized by nuclear area, , nucleolar eccentricity, , angle between the major nuclear and nucleolar axes, , the shortest distance from the nucleolar centroid to the nuclear envelope normalized by the nuclear circumference, , fraction of the nucleolar contour with negative curvature, , and number of continuous regions of negative curvature along the nucleolar contour, . All data collected in the nucleolar focal plane. Red dot, solid red line and dotted red lines indicate the mean, median and quartiles, respectively. Table 1 provides -values for all measured data with respect to the control. The number of nucleoli and cells are as follows: control ( 648, 208), blebbistatin ( 399, 127), latrunculin A ( 307, 104), nocodazole ( 310, 106), -amanitin ( 268, 95), flavopiridol ( 309, 105), trichostatin A ( 278, 95), and cycloheximide at 30 min ( 291, 91), and cycloheximide at 6.5 hr ( 294, 105). Scale bar, 5 µm.
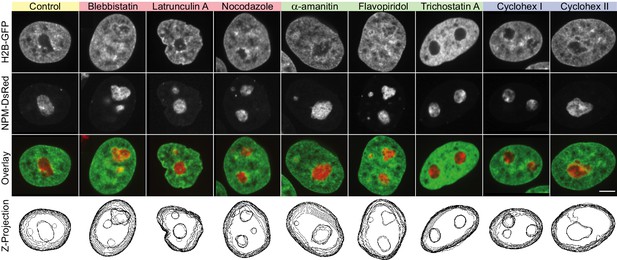
Nucleoli upon biochemical perturbations, including z-projections.
Micrographs of HeLa nuclei with fluorescently labeled chromatin (H2B-GFP, green) and nucleoli (NPM-DsRed, red), the two signal overlay, and z-projections of nuclear and nucleolar contours, under the following conditions: control, upon addition of blebbistatin, latrunculin A, nocodazole, -amanitin, flavopiridol, trichostatin A, and cycloheximide (at 30 min and 6.5 hr). Scale bar, 5 µm.
Tables
Statistical characteristics of distributions for physical quantities evaluated for nucleoli upon biochemical perturbations (see Figures 6–7).
| Mean ± standard deviation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 648 | 208 | 0.052 ± 0.040 | 1.30 ± 0.32 | 47 ± 26 | 0.076 ± 0.029 | 0.033 ± 0.066 | 0.66 ± 1.37 | ||||
| ATP-depletion | 345 | 127 | 0.054 ± 0.042 | 1.29 ± 0.28 | 48 ± 26 | 0.082 ± 0.028 | 0.038 ± 0.064 | 0.82 ± 1.39 | ||||
| Blebbistatin | 399 | 127 | 0.052 ± 0.041 | 1.29 ± 0.28 | 46 ± 26 | 0.076 ± 0.028 | 0.036 ± 0.067 | 0.72 ± 1.38 | ||||
| Latrunculin A | 307 | 104 | 0.061 ± 0.044 | 1.31 ± 0.36 | 47 ± 25 | 0.071 ± 0.028 | 0.030 ± 0.063 | 0.56 ± 1.21 | ||||
| Nocodazole | 310 | 106 | 0.052 ± 0.037 | 1.28 ± 0.25 | 50 ± 28 | 0.074 ± 0.029 | 0.032 ± 0.061 | 0.67 ± 1.38 | ||||
| -amanitin | 268 | 95 | 0.060 ± 0.047 | 1.34 ± 0.29 | 44 ± 27 | 0.074 ± 0.029 | 0.046 ± 0.072 | 0.99 ± 1.71 | ||||
| Flavopiridol | 309 | 105 | 0.052 ± 0.040 | 1.34 ± 0.34 | 43 ± 27 | 0.074 ± 0.028 | 0.048 ± 0.080 | 0.92 ± 1.67 | ||||
| Trichostatin A | 278 | 95 | 0.044 ± 0.032 | 1.29 ± 0.36 | 43 ± 28 | 0.073 ± 0.026 | 0.025 ± 0.063 | 0.53 ± 1.35 | ||||
| Cyclohex I | 291 | 91 | 0.053 ± 0.037 | 1.32 ± 0.29 | 46 ± 26 | 0.076 ± 0.025 | 0.038 ± 0.069 | 0.76 ± 1.49 | ||||
| Cyclohex II | 294 | 105 | 0.052 ± 0.039 | 1.35 ± 0.38 | 47 ± 25 | 0.074 ± 0.029 | 0.040 ± 0.067 | 0.74 ± 1.33 | ||||
| p-values (with respect to control) | Relative difference of mean [%] | |||||||||||
| ATP-depletion | 0.359 | 0.674 | 0.513 | 0.009 | 0.258 | 0.093 | 4% | −1% | 2% | 8% | 15% | 24% |
| Blebbistatin | 0.893 | 0.515 | 0.694 | 0.911 | 0.582 | 0.495 | 0% | −1% | −2% | 0% | 9% | 9% |
| Latrunculin A | 0.002 | 0.714 | 0.940 | 0.011 | 0.464 | 0.246 | 17% | 1% | 0% | −7% | −9% | −15% |
| Nocodazole | 0.985 | 0.376 | 0.169 | 0.335 | 0.734 | 0.925 | 0% | −2% | 6% | −3% | −3% | 2% |
| -amanitin | 0.011 | 0.089 | 0.090 | 0.280 | 0.017 | 0.006 | 15% | 3% | −6% | −3% | 39% | 50% |
| Flavopiridol | 0.789 | 0.089 | 0.059 | 0.272 | 0.006 | 0 .018 | 0% | 3% | −9% | −3% | 45% | 39% |
| Trichostatin A | 0.002 | 0.906 | 0.051 | 0.099 | 0.087 | 0.160 | −15% | −1% | −9% | −4% | −24% | −20% |
| Cyclohex I | 0.654 | 0.257 | 0.633 | 0.932 | 0.355 | 0.359 | 2% | 2% | −2% | 0% | 15% | 15% |
| Cyclohex II | 0.984 | 0.035 | 0.982 | 0.284 | 0.163 | 0.381 | 0% | 4% | 0% | −3% | 21% | 12% |
| Kullback-Leibler divergence (with respect to control) | Skew | |||||||||||
| Control | – | – | – | – | – | – | 1.10 | 3.07 | −0.09 | −0.04 | 2.17 | 2.68 |
| ATP-depletion | 0.026 | 0.011 | 0.021 | 0.032 | 0.051 | 0.018 | 1.01 | 2.23 | −0.19 | −0.16 | 1.80 | 1.87 |
| Blebbistatin | 0.023 | 0.025 | 0.031 | 0.036 | 0.022 | 0.007 | 1.31 | 2.26 | −0.11 | 0.03 | 2.00 | 2.14 |
| Latrunculin A | 0.059 | 0.029 | 0.028 | 0.040 | 0.050 | 0.015 | 0.79 | 3.01 | −0.15 | −0.07 | 2.06 | 2.36 |
| Nocodazole | 0.022 | 0.014 | 0.042 | 0.025 | 0.033 | 0.010 | 0.87 | 1.84 | −0.28 | 0.07 | 1.90 | 2.55 |
| -amanitin | 0.060 | 0.043 | 0.057 | 0.032 | 0.062 | 0.036 | 0.98 | 1.95 | 0.08 | 0.08 | 1.61 | 2.26 |
| Flavopiridol | 0.038 | 0.027 | 0.044 | 0.035 | 0.053 | 0.021 | 1.28 | 2.36 | 0.03 | −0.02 | 1.62 | 2.09 |
| Trichostatin A | 0.056 | 0.046 | 0.062 | 0.059 | 0.047 | 0.034 | 0.85 | 3.15 | 0.17 | −0.25 | 2.95 | 3.15 |
| Cyclohex I | 0.041 | 0.032 | 0.043 | 0.051 | 0.043 | 0.015 | 1.13 | 2.13 | −0.08 | −0.12 | 1.98 | 2.70 |
| Cyclohex II | 0.013 | 0.026 | 0.049 | 0.021 | 0.040 | 0.014 | 0.98 | 4.19 | −0.08 | 0.01 | 1.66 | 2.14 |
-
Table 1—source data 1
Statistical characteristics of distributions measured in Figures 6 and 7.
- https://cdn.elifesciences.org/articles/47533/elife-47533-table1-data1-v1.xlsx
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (H. sapiens) | HeLa | ATCC | ATCC:CCL2 RRID:CVCL_0030 | Stable H2B-GFP cell line |
| Transfected construct (H. sapiens) | NPM-DsRed | Addgene | 34553 RRID:Addgene_34553 | |
| Transfected construct (H. sapiens) | mCerulean-Fibrillarin-7 | Addgene | 55368 RRID:Addgene_55368 | |
| Transfected construct (H. sapiens) | NPM-mApple | Caragine et al., 2018 | n/a | Generated by Zidovska Lab – published in Caragine et al., 2018 |
| Chemical compound, drug | RO-3306 | Enzo Life Sciences | ALX-270–463 | |
| Chemical compound, drug | 2-Deoxy-D-Glucose (DOG) | Millipore Sigma | D8375 | |
| Chemical compound, drug | Trifluoromethoxy-carbonylcyanide phenylhydrazone (FCCP) | Millipore Sigma | C2920 | |
| Chemical compound, drug | Latrunculin A | Millipore Sigma | L5163 | |
| Chemical compound, drug | Blebbistatin | Millipore Sigma | B0560 | |
| Chemical compound, drug | Nocodazole | Millipore Sigma | M1404 | |
| Chemical compound, drug | α-Amanitin | Santa Cruz Biotechnology | sc-202440 | |
| Chemical compound, drug | Cycloheximide | Santa CruzBiotechnology | sc-3508 | |
| Chemical compound, drug | Flavopiridol | Santa Cruz Biotechnology | sc-202157 | |
| Chemical compound, drug | Trichostatin A | Millipore Sigma | T8552 | |
| Software, algorithm | Matlab | MathWorks | 2017a, 2019a | |
| Software, algorithm | Adobe Illustrator, Photoshop | Adobe Inc. | CC2018 |





