Mechanisms of chromosome biorientation and bipolar spindle assembly analyzed by computational modeling
Figures
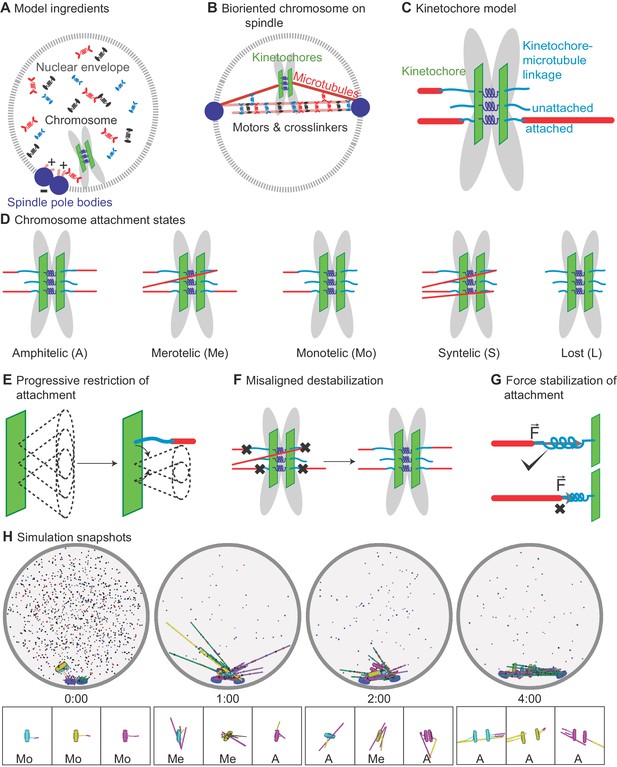
Schematic of computational model and simulation of the reference model.
(A) Schematic of initial condition, showing adjacent spindle-pole bodies (blue) embedded in the nuclear envelope (gray dashed), proximal chromosomes (gray with green plate and blue springs), short microtubules (pink), and motor proteins and crosslinkers (red, blue, and black). (B) Schematic of bipolar spindle and a bioriented chromosome. (C) Schematic of chromosome and kinetochore model showing sister chromatids (gray), one kinetochore on each chromatid (green plates), the pericentric chromatin spring (blue springs), and kinetochore-MT attachment factor (blue line). (D) Schematic of chromosome attachment states, showing amphitelic, merotelic, monotelic, syntelic, and lost chromosomes. (E) Schematic of progressive restriction, showing that the angular range of kinetochore-MT attachment is restricted after attachment. (F) Schematic of misaligned destabilization of attachment, showing that misaligned attachments are destabilized. (G) Schematic of force stabilization of attachment, showing that end-on attachment to depolymerizing MTs has increased lifetime. (H) Image sequence of spindle assembly and chromosome biorientation rendered from a three-dimensional simulation. Initially, spindle-pole bodies (SPBs) are adjacent (blue disks), MTs are short spherocylinders (green and purple when unattached to kinetochores, yellow and magenta when attached), and chromosomes (cyan, yellow, magenta) are near SPBs. Motors and crosslinkers are dispersed spots (red, blue, and black) within the nucleus (gray boundary). Time shown in minutes:seconds. Lower: a zoomed view of each chromosome with attachment state labeled.
-
Figure 1—source data 1
Configuration files for the simulations used for snapshots in Figure 1H.
- https://cdn.elifesciences.org/articles/48787/elife-48787-fig1-data1-v2.gz.zip
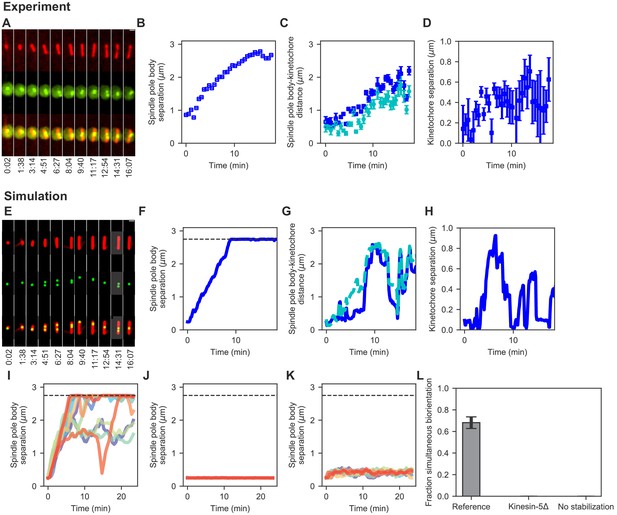
Comparison of spindle assembly and chromosome alignment in cells and simulations.
(A–D) Experimental results. (A) Maximum-intensity-projected smoothed images from time-lapse confocal fluorescence microscopy of fission yeast with mCherry-atb2 labeling MTs (red) and cen2-GFP labeling the centromere of chromosome 2 (green). Time shown in minutes:seconds. (B) Spindle length, (C) spindle pole body-kinetochore distance, and (D) interkinetochore distance versus time for the experiment shown in (A). (E–K) Simulation results. (E) Simulated fluorescence microscopy images with MTs (red) and a single kinetochore pair (green). (F) Spindle length, (G) spindle pole body-kinetochore distance, and (H) interkinetochore distance versus time from the simulation shown in (E), sampled at a rate comparable to the experimental data in (A–D). Note that the rigid nucleus in our model sets an upper limit on spindle length of 2.75 μm, as shown by the dashed line in F. (I) Spindle length versus time for 12 simulations of the reference model. (J) Spindle length versus time for 12 simulations in a model lacking kinesin-5. (K) Spindle length versus time for 12 simulations in a model lacking crosslink-mediated microtubule stabilization. (L) Fraction of simultaneous biorientation for the reference, kinesin-5 delete, and no-stabilization models (N = 12 simulations per data point).
-
Figure 2—source data 1
Configuration and data files for the simulations used in Figure 2.
Configuration files are contained within *config.tar.gz. Data are contained within *data.csv files.
- https://cdn.elifesciences.org/articles/48787/elife-48787-fig2-data1-v2.gz.zip
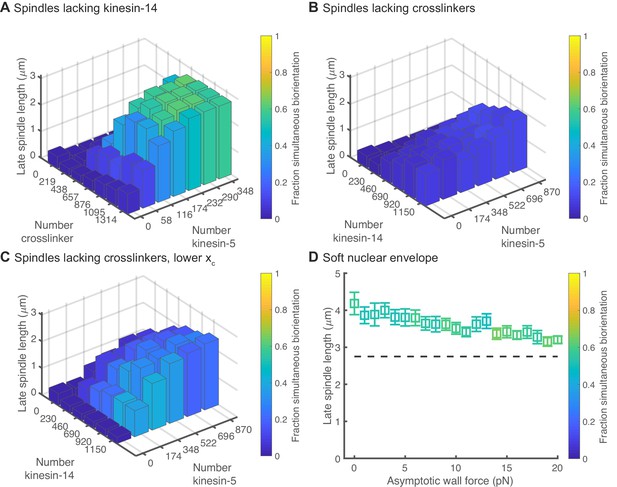
Results of simulations with perturbations to motor and crosslinker number, motor force-dependent unbinding, and nuclear envelope rigidity.
(A) Simulated spindles form and biorient chromosomes in the absence of kinesin-14 motors if kinesin-5 and crosslinker number are increased. (B) Simulated spindles have difficulty forming in the absence of crosslinkers, and do not properly biorient chromosomes. (C) Lowering the characteristic distance of force-dependent unbinding of kinesin-14 to that of kinesin-5 (which makes kinesin-14 motors less sensitive to force-induced unbinding) causes longer spindles to form that are capable of biorienting chromosomes. (D) Spindle length as a function of wall force for a model of a soft nuclear envelope for which the SPBs are not fixed on the surface of the sphere. The reference model contains 174 kinesin-5 motors, 230 kinesin-14 motors, and 657 crosslinkers. (N = 12 simulations per data point.).
-
Figure 2—figure supplement 1—source data 1
Configuration and data files for simulations used in Figure 2—figure supplement 1.
Configuration files are contained within the *config.tar.gz files. Data are contained within the MATLAB scripts *panelA-D.m.
- https://cdn.elifesciences.org/articles/48787/elife-48787-fig2-figsupp1-data1-v2.gz.zip
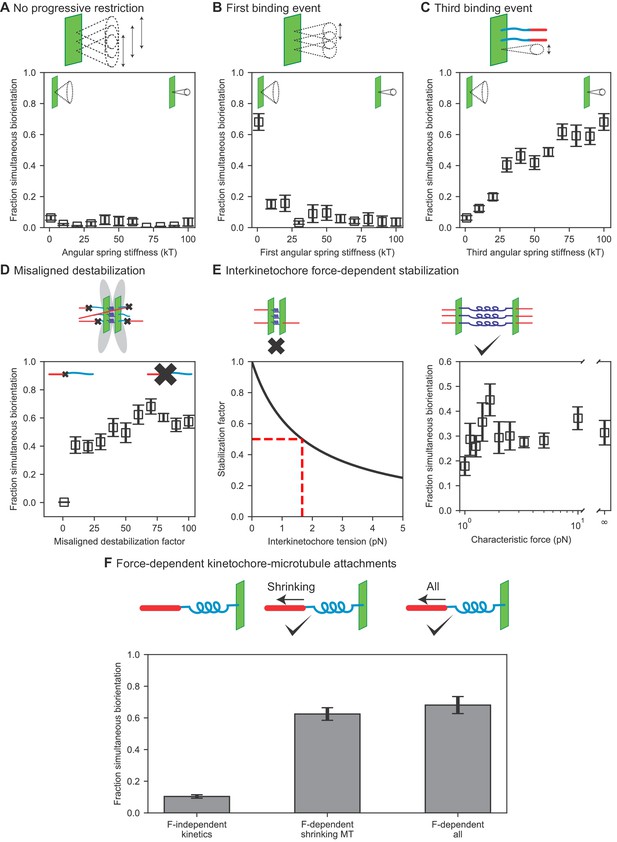
Results of perturbing kinetochore properties required for biorientation.
(A) Fraction simultaneous biorientation versus angular spring stiffness in models lacking progressive restriction of attachment. (B) Fraction simultaneous biorientation versus the first angular spring stiffness in the model with progressive restriction. (C) Fraction simultaneous biorientation versus the third angular spring stiffness in the model with progressive restriction. (D) Fraction simultaneous biorientation versus the misaligned destabilization factor. (E) Effects of force-dependent error correction. Top, schematic of stabilization of kinetochore-MT attachments as a function of interkinetochore force. Left, Stabilization as a function of interkinetochore tension for a characteristic force of 1.67 pN. When the interkinetochore force is the characteristic force, attachment turnover is reduced by a factor of two, as shown by the red dashed lines. Right, fraction simultaneous biorientation versus the characteristic force. (F) Fraction simultaneous biorientation for different types of force-dependent kinetics (N = 12 simulations per data point).
-
Figure 3—source data 1
Configuration and data files for simulations used in Figure 3.
- https://cdn.elifesciences.org/articles/48787/elife-48787-fig3-data1-v2.gz.zip
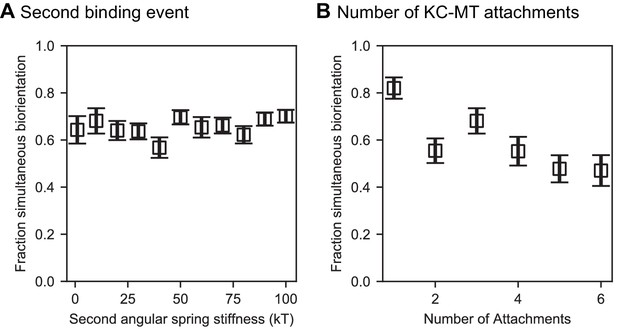
Effects of varying the middle angular stiffness for progressive restriction and the number of kinetochore-microtubule attachments.
(A) Angular spring stiffness for the middle value chosen in progressive restriction did not affect chromosome biorientation fidelity. In these models, the first and third angular spring stiffnesses were fixed at 1 and 100 , respectively. (B) Varying the number of microtubule attachment sites per kinetochore does not significantly alter biorientation in the model. We varied the angular spring stiffnesses are varied with the number of attachments shown as (1: [1 , 1 ], 2: [1 , 10 , 10 ], 3: Reference model, 4–6: [1 , 10 , 100 , 100 , … 100 ]) (N = 12 simulations per data point).
-
Figure 3—figure supplement 1—source data 1
Configuration and data files for simulations used in Figure 3—figure supplement 1.
Configuration files are contained within the *config.tar.gz files. Data are contained within the *data.csv files.
- https://cdn.elifesciences.org/articles/48787/elife-48787-fig3-figsupp1-data1-v2.gz.zip
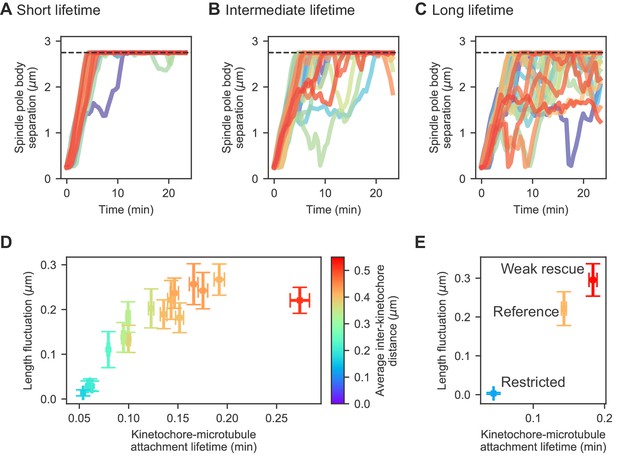
Changes in kinetochore-MT attachment turnover alter spindle length fluctuations.
(A–C) Spindle length versus time for 24 simulations of the same model, with (A) short (1/4 the reference value), (B) intermediate (1/2 the reference value), and (C) long (twice the reference value) kinetochore-MT attachment lifetime. (D) Length fluctuation magnitude versus measured kinetochore-MT attachment lifetime and average interkinetochore stretch (color) for bioplar spindles (corresponding to simulation time >10 min.). (E) Length fluctuation magnitude versus measured kinetochore-MT attachment lifetime and average interkinetochore stretch (color) for the reference, restricted, and weak rescue models (N = 24 simulations per data point).
-
Figure 4—source data 1
Configuration and data files for simulations used in Figure 4.
Configuration files are contained within *config.tar.gz files for the noted panels. Figure 4A–C data contained within *data.csv files that pertains to the spindle length versus time measurements. Data for panels Figure 4D,E are contained in their respective files as well, but contain data for the kinetochore-microtubule attachment lifetimes and length fluctuations measurements.
- https://cdn.elifesciences.org/articles/48787/elife-48787-fig4-data1-v2.gz.zip
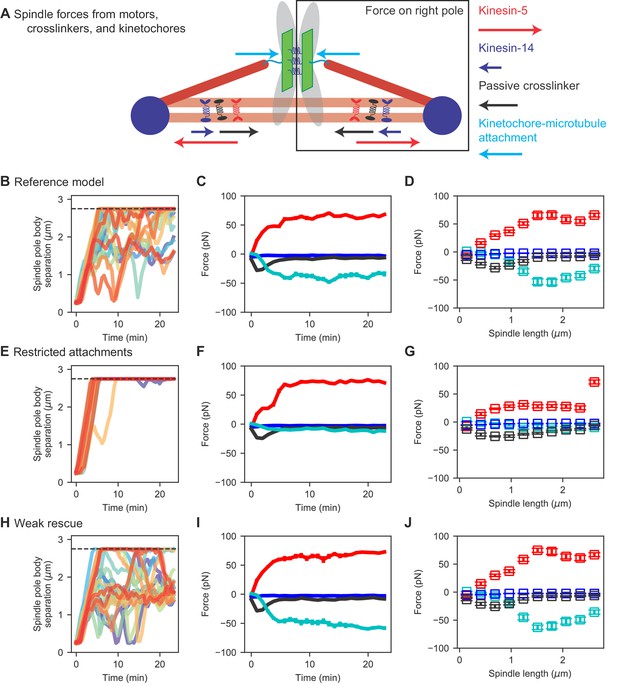
Spindle force generation varies as the spindle assembles and elongates.
(A) Schematic of force generation along the spindle axis, showing kinesin-5 motors exerting outward force (red) and kinesin-14 (dark blue), crosslinkers (black), and kinetochore-MT attachment to stretched chromosomes (light blue) exerting inward force. (B, E, H) Spindle length versus time, (C, F, I) average spindle axis force versus time, and (D, G, J) average spindle axis force versus spindle length for three different models: (B–D) the reference model, (E–G) the restricted attachment model, and (H–J) the weak rescue model (N = 24 simulations per data point).
-
Figure 5—source data 1
Configuration and data files for simulations used in Figure 5.
Configuration files are contained within *config.tar.gz files for the noted panels. Spindle length versus time measurements are found within Figure 5b,e,h_data.csv files. Spindle force measurements are found within Figure 5cd,fg,ij_data.csv files.
- https://cdn.elifesciences.org/articles/48787/elife-48787-fig5-data1-v2.gz.zip
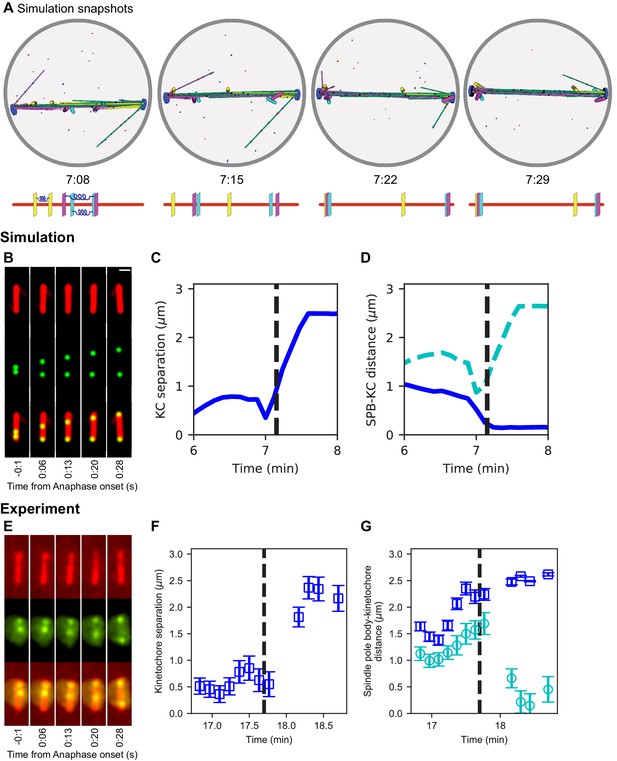
Chromosome segregation in the model and comparison to experiments.
(A) Image sequence of simulation of chromosome segregation after anaphase is triggered, rendered from a three-dimensional simulation. Anaphase begins immediately after the first image. Lower, schematic showing kinetochore position along the spindle. Time shown in minutes:seconds. (B–D) Simulation results. (B) Simulated fluorescence microscopy images with MTs (red) and a single kinetochore pair (green). Time shown in minutes:seconds. (C) Spindle pole body-kinetochore distance, and (D) interkinetochore distance versus time from the simulation shown in (B), sampled at a rate comparable to the experimental data in (E–G). (E–G) Experimental results. Maximum-intensity projected smoothed images from time-lapse confocal fluorescence microscopy of fission yeast with mCherry-atb2 labeling MTs (red) and cen2-GFP labeling the centromere of chromosome 2 (green). Time shown in minutes:seconds. (E) Spindle length, (F) spindle pole body-kinetochore distance, and (G) interkinetochore distance versus time from the experiment shown in (E).
-
Figure 6—source data 1
Configuration and data files for simulations used in Figure 6.
Configuration files are contained within *config.tar.gz files for the noted panels. Spindle length data are found within the *data.csv files.
- https://cdn.elifesciences.org/articles/48787/elife-48787-fig6-data1-v2.gz.zip
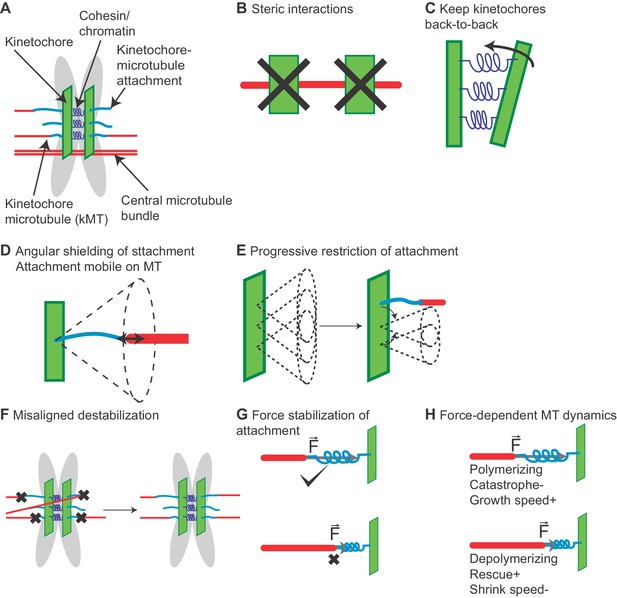
Chromosome model overview.
(A) Chromosomes are modeled as sister chromatids and kinetochores held together by a cohesin/chromatin spring complex. Each kinetochore can attach up to three microtubules. (B) Steric interactions between MTs and kinetochores prevent overlap, while a soft steric repulsion exists between MTs and the centromeric DNA. (C) Kinetochores are kept back-to-back through a cohesin-chromatin spring complex that depends on relative kinetochore position and orientation. (D) The angular range of kinetochore-MT attachment is restricted based on the stiffness of an angular spring. (E) The angular restriction of kinetochore-MT attachment changes based on the number of bound MTs. (F) Attachments are destabilized when the chromosome is not properly bioriented. (G) Attachment lifetime is force-dependent, with attachments to depolymerizing MTs under tension having longer lifetimes, while those to polymerizing MTs have their lifetime decreased under tension. (H) MT dynamics are force-dependent. Polymerizing MTs have increased growth speed and reduced catastrophe, while depolymerizing MTs have increased rescue and decreased shrinking speed.
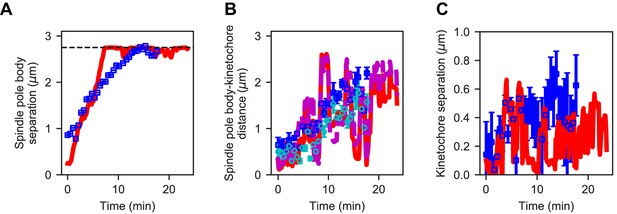
Reference model generates similar dynamics of spindle length and kinetochore position compared to experiment.
(A) Spindle length versus time for experiment (blue) and refined model (red). (B) Spindle pole body-kinetochore distance versus time for a single kinetochore pair (Cen2) in experiment (blue, cyan) and refined model (red, magenta). (C) Kinetochore separation versus time for experiment (blue) and refined model (red). This comparison gives Pearson correlation coefficients for length = 0.891, SPB-KC distance = 0.72, Interkinetochore distance = 0.42.
Videos
Simulation of the reference model shows spindle assembly simultaneous with chromosome biorientation.
Initially, short MTs begin to grow at the start of the simulation and interact with nearby kinetochores. A bipolar spindle forms as the chromosomes begin to biorient. Finally, a metaphase spindle is established with bioriented chromosomes that move along the spindle and breathe. The insets are zoomed views of each chromosome, showing attachment turnover and interkinetochore stretch.
Top: Simulation of reference model (left) and simulated fluorescence microscopy images (right), with red MTs and green kinetochore (scale bar 1 μm).
The simulated fluorescence images are rotated so that the spindle is vertical. Lower: simulation of models mimicking genetic perturbation. Lower left: Model lacking kinesin-5 motors. The SPBs never separate and the spindle remains monopolar. Chromosomes do not biorient. Lower right: Model lacking crosslinker-mediated stabilization of MT dynamics. SPBs separate only slightly, forming a short spindle that is nearly indistinguishable from a monopolar spindle. Chromosomes do not biorient.
Simulation of a model with a soft nuclear envelope and an asymptotic wall force on the SPBs of 17 pN.
SPBs are able to move away from their preferred radius from the center of the nucleus. The spindle reaches a bounded length, and chromosomes are able to biorient. Spindle length larger than the nuclear envelope radius is reached by the balance of force from motors, crosslinkers, chromosomes.
Simulations of models with perturbation to kinetochore properties important for biorientation.
Top left: Model lacking progressive restriction, with a common angular spring stiffnesses of 1 for all attachments. A short bipolar spindle forms, but chromosomes are typically merotelically attached and do not biorient. Top middle: Model lacking progressive restriction, with a common angular spring stiffnesses of 100 for all attachments. A long bipolar spindle forms, kinetochore-MT attachments are transient, and chromosomes do not generate significant inward force on the spindle. Top right: Model including progressive restriction with an angular spring stiffness of 20 for the first binding event, leading to restricted attachments. A long bipolar spindle forms, and kinetochore-MT attachments are transient. Lower left: model including progressive restriction but with an angular spring stiffness of 20 for the third binding event, leading to permissive attachments. Error correction is impaired, and chromosomes are typically merotelically attached. Lower middle: Model lacking misaligned destabilization. Error correction is impaired. Lower right: Model with force-independent attachment kinetics. Kinetochore-MT attachments are not stabilized under tension from depolymerizing microtubules, leading to short-lived biorientation.
Simulation of a model with interkinetochore force-dependent attachments.
The spindle forms in a few minutes, and chromosomes form stable, bioriented attachments. Zoomed views of chromosomes shows them forming load-bearing attachments to the tips of MTs. The interkinetochore characteristic force is 1.67 pN.
Simulations of models with varying kinetochore-MT attachment lifetime.
Left: Model with short attachment lifetime in which the kinetochore-MT binding and unbinding rates are 4 times larger than in the reference model. Biorientation is somewhat compromised. Middle: Model with intermediate attachment lifetime in which the kinetochore-MT binding and unbinding rates are 2 times larger than in the reference model. Right: Model with long attachment lifetime in which the kinetochore-MT binding and unbinding rates are 2 times smaller than in the reference model. Biorientation is preserved and the spindle undergoes large length fluctuations.
Simulations of reference, restricted, and weak rescue models.
Left: The reference model shows typical spindle length fluctuations. Middle: The restricted attachment model shows minimal length fluctuations, because transient kinetochore-MT attachments lead to low inward force on the spindle from chromosomes. Right: The weak rescue model shows large spindle length fluctuations, because kinetochore MTs remain attached while depolymerizing, leading to high and fluctuating inward force on the spindle from chromosomes.
Simulations of anaphase chromosome segregation.
Top: Simulation video showing that separation of the sister chromatids occurs after 4.45 min of the simultaneous biorientation of all three chromosomes. The zoomed views show the chromosomes achieving biorientation before segregating to the spindle poles. Lower: Simulation video (left) and simulated fluorescence microscopy images (right), with red MTs and green kinetochore (scale bar 1 μm). The simulated fluorescence images are rotated so that the spindle is vertical. Anaphase occurs at 7:09.
Tables
Simulation, SPB, and MT parameters.
| Simulation parameter | Symbol | Value | Notes |
|---|---|---|---|
| Time step | 8.9 ×10-6 s | Blackwell et al., 2017a | |
| Nuclear envelope radius | 1.375 μm | Kalinina et al., 2013 | |
| Spindle pole bodies | |||
| Diameter | 0.1625 μm | Ding et al., 1993 | |
| Bridge size | 75 nm | Ding et al., 1993 | |
| Tether length | 50 nm | Flory et al., 2002; Muller et al., 2005 | |
| Tether spring constant | 0.6625 pN nm-1 | Blackwell et al., 2017a | |
| Translational diffusion coefficient | 4.5 × 10-4 μm2 s-1 | Blackwell et al., 2017a | |
| Rotational diffusion coefficient | 0.0170 s-1 | Blackwell et al., 2017a | |
| Linkage time | 5 s | Blackwell et al., 2017a | |
| Microtubules | |||
| Diameter | 25 nm | Blackwell et al., 2017a | |
| Angular diffusion coefficient | Depends on MT length | Blackwell et al., 2017a; Kalinina et al., 2013 | |
| Force-induced catastrophe constant | 0.5 pN-1 | Blackwell et al., 2017a; Janson et al., 2003; Dogterom and Yurke, 1997 | |
| Growth speed | 4.1 μm min-1 | Blackwell et al., 2017a; Blackwell et al., 2017b | |
| Shrinking speed | 6.7 μm min-1 | Blackwell et al., 2017a; Blackwell et al., 2017b | |
| Catastrophe frequency | 3.994 min-1 | Blackwell et al., 2017a; Blackwell et al., 2017b | |
| Rescue frequency | 0.157 min-1 | Blackwell et al., 2017a; Blackwell et al., 2017b | |
| Growth speed stabilization | 1.54 | Optimized | |
| Shrinking speed stabilization | 0.094 | Optimized | |
| Catastrophe frequency stabilization | 0.098 | Optimized | |
| Rescue frequency stabilization | 18 | Optimized | |
| Stabilization length | 16 nm | Optimized | |
| Minimum MT length | 75 nm | Optimized | |
Soft nuclear envelope model parameters.
| Parameter | Symbol | Value | Notes |
|---|---|---|---|
| Translational mobility | Calculated | ||
| Rotational mobility | Calculated | ||
| Membrane tube radius | 87.7 nm | Derényi et al., 2002; Lim et al., 2007; Lamson et al., 2019 | |
| MT asymptotic wall force | 2.5 pN | Derényi et al., 2002; Lim et al., 2007; Lamson et al., 2019 | |
| SPB asymptotic wall force | 17 pN | Derényi et al., 2002; Lim et al., 2007; Lamson et al., 2019 | |
| Tether spring constant | 6.625 pN nm-1 | Optimized |
Motor and crosslinker parameters.
| Simulation parameter | Symbol | Value | Notes |
|---|---|---|---|
| Kinesin-5 | |||
| Number | 174 | Optimized (Carpy et al., 2014) | |
| Association constant per site | 90.9 μM-1 site-1 | Cochran et al., 2004 | |
| One-dimensional effective concentration | 0.4 nm-1 | Blackwell et al., 2017a | |
| Spring constant | Kawaguchi and Ishiwata, 2001 | ||
| Singly-bound velocity | Roostalu et al., 2011 | ||
| Polar aligned velocity | Gerson-Gurwitz et al., 2011 | ||
| Anti-polar aligned velocity | Gerson-Gurwitz et al., 2011 | ||
| Singly bound off-rate | 0.11 s-1 | Roostalu et al., 2011 | |
| Doubly bound off-rate (single head) | 0.055 s-1 | Blackwell et al., 2017a | |
| Tether length | 53 nm | Kashlna et al., 1996 | |
| Stall force | 5 pN | Valentine et al., 2006 | |
| Characteristic distance | 1.5 nm | Optimized (Arpağ et al., 2014 | |
| Diffusion constant (solution) | 4.5 | Bancaud et al., 2009 | |
| Kinesin-14 | |||
| Number | 230 | Optimized (Carpy et al., 2014) | |
| Association constant (motor head) | 22.727 μM-1 site-1 | Chen et al., 2012 | |
| Association constant (passive head) | 22.727 μM-1 site-1 | Blackwell et al., 2017a | |
| 1D effective concentration (motor head) | 0.1 nm-1 | Blackwell et al., 2017a | |
| 1D effective concentration (passive head) | 0.1 nm-1 | Blackwell et al., 2017a | |
| Spring constant | Kawaguchi and Ishiwata, 2001 | ||
| Singly bound velocity (motor head) | Blackwell et al., 2017a | ||
| Diffusion constant (bound, diffusing head) | 0.1 μm2 s-1 | Blackwell et al., 2017a | |
| Singly bound off-rate (motor head) | 0.11 s-1 | Blackwell et al., 2017a | |
| Singly bound off-rate (passive head) | 0.1 s-1 | Blackwell et al., 2017a | |
| Doubly bound off-rate (motor head) | 0.055 s-1 | Blackwell et al., 2017a | |
| Doubly bound off-rate (passive head) | 0.05 s-1 | Blackwell et al., 2017a | |
| Tether length | 53 nm | Blackwell et al., 2017a | |
| Stall force | 5.0 pN | Blackwell et al., 2017a | |
| Characteristic distance | 4.8 nm | Optimized (Arpağ et al., 2014) | |
| Adjusted characteristic distance | 1.5 nm | Figure 2—figure supplement 1C | |
| Crosslinker | |||
| Number | 657 | Optimized (Carpy et al., 2014) | |
| Association constant | 90.9 μM-1 site-1 | Cochran et al., 2004 | |
| One-dimensional effective concentration | 0.4 nm-1 | Lansky et al., 2015 | |
| Spring constant | 0.207 pN nm-1 | Lansky et al., 2015 | |
| Diffusion constant (solution) | 4.5 | Bancaud et al., 2009 | |
| Singly bound diffusion constant | Lansky et al., 2015 | ||
| Doubly bound diffusion constant | Lansky et al., 2015 | ||
| Singly bound off-rate | 0.1 s-1 | Kapitein et al., 2008 | |
| Doubly bound off-rate | 0.05 s-1 | Lansky et al., 2015 | |
| Parallel-to-antiparallel bindng ratio | 0.33 | Kapitein et al., 2008; Rincon et al., 2017; Lamson et al., 2019 | |
| Characteristic distance | 2.1 nm | Optimized (Arpağ et al., 2014) | |
| Tether length | 53 nm | Lansky et al., 2015; Lamson et al., 2019 | |
Chromosome and kinetochore parameters.
| Simulation parameter | Symbol | Value | Notes |
|---|---|---|---|
| Kinetochore kinematics | |||
| Diameter | 200 nm | Blackwell et al., 2017a; Kalinina et al., 2013 | |
| Length | 150 nm | Ding et al., 1993 | |
| Width | 50 nm | Ding et al., 1993 | |
| Thickness | 0 nm | Chosen | |
| Diffusion coefficient | 5.9 × 10-4µm2 s-1 | Gergely et al., 2016; Blackwell et al., 2017a; Kalinina et al., 2013 | |
| Translational drag | 3.51 pN µm-1 s | Computed | |
| Rotational drag | 0.165 pN µm s | Computed | |
| Catastrophe enhancement | 0.5 pN-1 | Matches NE factor | |
| MT tip length | 25 nm | Chosen | |
| Interkinetochore spring | |||
| Tether length | 100 nm | Stephens et al., 2013; Gergely et al., 2016; Gay et al., 2012 | |
| Linear spring constant | 39 pN µm-1 | Optimized | |
| Rotational spring constant | 1850 pN nm rad-1 | Optimized | |
| Alignment spring constant | 1850 pN nm rad-1 | Optimized | |
| Pericentric chromatin | |||
| Pericentric chromatin length | 200 nm | Chosen | |
| Pericentric chromatin diameter | 75 nm | Chosen | |
| Kinetochore-centromere offset | 37.5 nm | Chosen | |
| Chromatin-MT repulsion amplitude | 1 pN nm | Optimized | |
Attachment factor parameters.
| Parameter | Symbol | Value | Notes |
|---|---|---|---|
| Number | 3 | Ding et al., 1993 | |
| Attachment-site separation on kinetochore | 40 nm | Ding et al., 1993 | |
| Linear spring constant | 0.088 pN nm-1 | Optimized | |
| Angular spring constant, 0 to 1 | 4.1 pN nm | Optimized | |
| Angular spring constant, 1 to 2 | 41 pN nm | Optimized | |
| Angular spring constant, 2 to 3 | 410 pN nm | Optimized | |
| Angular spring constant, 3 to 3 | 410 pN nm | Optimized | |
| Tether length | 54 nm | Ciferri et al., 2007 | |
| kMC steps | 10 | Chosen | |
| MT tip length | 25 nm | Chosen | |
| MT tip crowding | True | Ding et al., 1993 | |
| Tip concentration | 40 nm-1 | Optimized | |
| Side concentration | 0.4 nm-1 | Optimized | |
| Tip rate assembling | 0.0001 s-1 | Optimized | |
| Tip rate disassembling | 0.03 s-1 | Optimized | |
| Side rate | 0.03 s-1 | Optimized | |
| Tip characteristic distance assembling | 1 nm | Optimized | |
| Tip characteristic distance disassembling | −3.9 nm | Optimized | |
| Side characteristic distance | −0.37 nm | Optimized | |
| Angular characteristic factor | 0.013 | Optimized | |
| Speed | 50 nm s-1 | Optimized | |
| Stall force | 5 pN | Kinesin-5 (Blackwell et al., 2017a; Akera et al., 2015) | |
| Tip diffusion | 0.0012 μm2 s-1 | Optimized | |
| Side diffusion | 0.018 μm2 s-1 | Optimized | |
| Tip tracking | 0.25 | Optimized | |
| Tip-enhanced catastrophe | 4 | Optimized | |
| Misaligned destabilization | 70 | Optimized | |
| Polymerization force factor | 8.4 pN | Akiyoshi et al., 2010; Gergely et al., 2016 | |
| Depolymerization force factor | −3.0 pN | Akiyoshi et al., 2010; Gergely et al., 2016 | |
| Catastrophe force factor | −2.3 pN | Akiyoshi et al., 2010; Gergely et al., 2016 | |
| Rescue force factor | 6.4 pN | Akiyoshi et al., 2010; Gergely et al., 2016 | |
| Maximum polymerization speed | 30 μm min-1 | Gergely et al., 2016 |
Force-dependent error correction model parameters.
| Parameter | Symbol | Value | Notes |
|---|---|---|---|
| Inter-kinetochore stabilization force | 1.67 pN | Optimized | |
| Rotational spring constant | 925 pN nm rad-1 | Optimized | |
| Alignment spring constant | 925 pN nm rad-1 | Optimized | |
| Angular characteristic factor | 0.08 | Optimized | |
| Side concentration | 0.32 nm-1 | Optimized | |
| Kinesin-5 number | 200 | Optimized |
Anaphase parameters.
| Anaphase | Symbol | Value | Notes |
|---|---|---|---|
| Integrated simultaneous biorientation time | 4.45 min | Chosen | |
| Anaphase attachment rate | 0.00007 s-1 | Chosen | |
| Anaphase MT depoly speed | 2.2 µm min-1 | Chosen |
Strain used in this study.
| Name | Genotype | Notes |
|---|---|---|
| MB 998 | cen2::kanr-ura4+-lacOp, his7+::lacI-GFP, z:adh15:mCherry-atb2:natMX6, leu1-32, h- | This study |
Additional files
-
Source code 1
Code for simulation and analysis framework for confined SPB simulations.
Requires C++ compiler, and GSL, GLEW, python2, python3, libyaml, FFTW, GLFW, xQuartz, freeGLUT, libpng, ffmpeg, pkg-config, and png++ libraries. Python libraries should include matplotlib, numpy, opencv-python, panda3d, pandas, PyYAML, and scipy for analysis framework. Used for all simulations except Figure 2—figure supplement 1, panel D: Soft nuclear envelope. Untar and unzip SourceCodeFile1.tar.gz, then use the accompanying Makefile and MakefileIncmk to compile on your system.
- https://cdn.elifesciences.org/articles/48787/elife-48787-code1-v2.tar.gz
-
Source code 2
Code for simulation and analysis framework for free SPB simulations.
Requires C++ compiler, and armadillo, GSL, GLEW, python2, python3, libyaml, FFTW, GLFW, xQuartz, freeGLUT, libpng, ffmpeg, pkg-config, and png++ libraries. Python libraries should include matplotlib, numpy, opencv-python, panda3d, pandas, PyYAML, and scipy for analysis framework. Used only for Figure 2—figure supplement 1, panel D: Soft nuclear envelope. Untar and unzip SourceCodeFile2.tar.gz, then use the accompanying Makefile and MakefileIncmk to compile on your system.
- https://cdn.elifesciences.org/articles/48787/elife-48787-code2-v2.tar.gz
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/48787/elife-48787-transrepform-v2.docx





