Protein Homeostasis: How chloroplasts protect themselves from unfolded proteins
Eukaryotic cells contain organelles that perform specific roles. The chloroplasts in algae and plants, for example, harvest the energy from sunlight to produce organic molecules via photosynthesis. An organelle must import proteins from the cytoplasm to perform its role: however, these proteins have to be unfolded when they are imported, so they must be rapidly processed and folded once they are inside the organelle. If proteins that are not correctly folded accumulate, they can become toxic, and must be detected and removed in order to protect the organelle from damage.
A quality control system consisting of small heat shock proteins, chaperones and proteases ensures that all proteins entering the chloroplast are properly folded. However, when algae are exposed to high levels of light, or when a protease called ClpP1 is missing, this quality control system can become compromised and overwhelmed by unfolded proteins. This causes the chloroplast to trigger an ‘unfolded protein response’ (UPR) that upregulates the production of various proteins in the nucleus, which then travel to the chloroplast to help it cope with this overload of unfolded proteins. Although the UPR is well-understood in other organelles, such as the ER and mitochondria, the UPR in chloroplasts has been described to a far lesser degree. For example, it remains unclear how the chloroplast monitors the level of unfolded proteins and communicates this information to the nucleus.
Now, in eLife, Silvia Ramundo of the University of California San Francisco (UCSF) and colleagues – including Karina Perlaza as first author – report how they used a genetic screen to discover the first signaling component of chloroplast unfolded protein response (cpUPR) in the algae Chlamydomonas reinhardti (Perlaza et al., 2019). To create the screen, the researchers used an inducible switch that blocks the production of the ClpP1 protease: this blocking causes unfolded proteins to accumulate inside the chloroplast, triggering the cpUPR (Figure 1; Ramundo et al., 2014; Ramundo and Rochaix, 2014). To monitor the response, algae were engineered to express a fluorescent reporter that was under the control of a nuclear expressed gene that only becomes upregulated when the cpUPR is switched on. Perlaza et al. then inserted random mutations into the genome of these engineered algae, and screened for mutants that were no longer displaying fluorescence despite having activated the cpUPR (Figure 1).
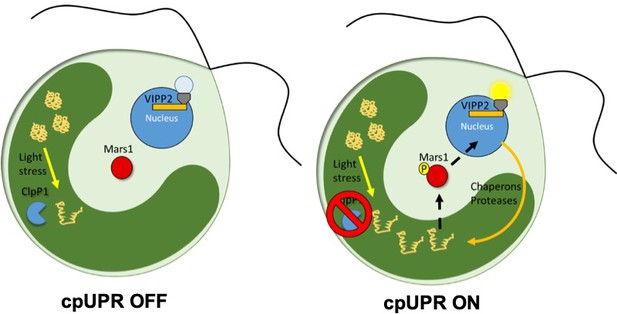
The unfolded protein response in chloroplasts.
In wild-type C. reinhardti algae a protease called ClpP1 is responsible for removing unfolded proteins produced by stress (left). The loss of ClpP1 (right) leads to the accumulation of unfolded proteins in the chloroplast (dark green) triggering the cpUPR. This causes nuclear genes which encode factors that assist in protein folding (such as chaperones and proteases) to become upregulated. In order to monitor changes to cpUPR signaling, a fluorescent protein was put under the control of one of these factors (VIPP2), so that fluorescence could only be observed when cpUPR was induced. By inserting random mutations into the genome and screening for mutants no longer displaying the fluorescent reporter, Perlaza et al. were able to identify MARS1, a kinase involved in transmitting the cpUPR signal from the chloroplast to the nucleus. MARS1: mutant affected in chloroplast-to-nucleus retrograde signaling 1.
Using this screen, the team – which includes researchers from UCSF, the Lawrence Berkeley Lab and Princeton – identified a gene coding for a protein called MARS1 that is essential for activating the cpUPR. Because MARS1 was localized outside of the chloroplast and contained a kinase domain (a critical component of cell signaling), Perlaza et al. deduced that MARS1 is likely to be involved in sending the cpUPR signal from the chloroplast to the nucleus. Furthermore, mutating the catalytic residue within the kinase domain revealed that kinase activity was critical for the cpUPR.
Stress caused by high levels of sunlight leads to an increased production of reactive oxygen species (ROS) that damage proteins inside the chloroplast, which in turn triggers the cpUPR. Perlaza et al. found that mutating the kinase domain of MARS1 caused algae to be more susceptible to this type of chloroplast stress. Notably, the increased susceptibility of the mutants was only linked to chloroplast stressors that trigger the cpUPR, suggesting that MARS1 is specific for the unfolded protein response. Perlaza et al. also observed that mutant algae that had a dominantly active version of the kinase displayed enhanced resistance to high levels of light and ROS. This opens a potential avenue for how the cpUPR could be exploited to increase the efficiency of photosynthesis in other organisms.
The study of cpUPR is still in its infancy, but it is already revealing its importance in chloroplast development and the protection of photosynthetic activity. A crucial next step will be to apply this knowledge to higher plants, where MARS1 has not been identified but where responses equivalent to cpUPR have been shown to exist (Llamas et al., 2017). Any new components of the cpUPR pathway that researchers are able to identify could be potential new targets for increasing yields in crop species.
References
-
Chloroplast unfolded protein response, a new plastid stress signaling pathway?Plant Signaling & Behavior 9:e972874.https://doi.org/10.4161/15592316.2014.972874
Article and author information
Author details
Publication history
- Version of Record published: October 15, 2019 (version 1)
Copyright
© 2019, Kessler and Longoni
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,861
- views
-
- 197
- downloads
-
- 6
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cell Biology
- Structural Biology and Molecular Biophysics
Mutations in the human PURA gene cause the neurodevelopmental PURA syndrome. In contrast to several other monogenetic disorders, almost all reported mutations in this nucleic acid-binding protein result in the full disease penetrance. In this study, we observed that patient mutations across PURA impair its previously reported co-localization with processing bodies. These mutations either destroyed the folding integrity, RNA binding, or dimerization of PURA. We also solved the crystal structures of the N- and C-terminal PUR domains of human PURA and combined them with molecular dynamics simulations and nuclear magnetic resonance measurements. The observed unusually high dynamics and structural promiscuity of PURA indicated that this protein is particularly susceptible to mutations impairing its structural integrity. It offers an explanation why even conservative mutations across PURA result in the full penetrance of symptoms in patients with PURA syndrome.
-
- Cell Biology
Endogenous tags have become invaluable tools to visualize and study native proteins in live cells. However, generating human cell lines carrying endogenous tags is difficult due to the low efficiency of homology-directed repair. Recently, an engineered split mNeonGreen protein was used to generate a large-scale endogenous tag library in HEK293 cells. Using split mNeonGreen for large-scale endogenous tagging in human iPSCs would open the door to studying protein function in healthy cells and across differentiated cell types. We engineered an iPS cell line to express the large fragment of the split mNeonGreen protein (mNG21-10) and showed that it enables fast and efficient endogenous tagging of proteins with the short fragment (mNG211). We also demonstrate that neural network-based image restoration enables live imaging studies of highly dynamic cellular processes such as cytokinesis in iPSCs. This work represents the first step towards a genome-wide endogenous tag library in human stem cells.






