Existence and functions of a kisspeptin neuropeptide signaling system in a non-chordate deuterostome species
Figures
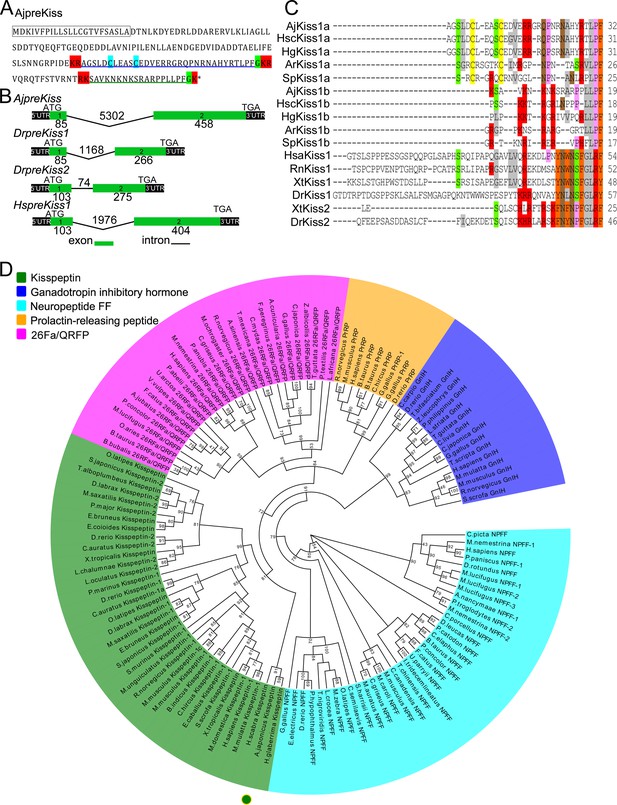
Gene structure, homology, and phylogenetic characterization of Apostichopus japonicus kisspeptin precursor (AjpreKiss).
(A) Deduced amino-acid sequence of AjpreKiss. The signal peptide is labeled in the box with full lines; the cleavage sites are highlighted in red; glycine residues responsible for C-terminal amidation are highlighted in green; cysteines paired in a disulfide-bonding structure are highlighted in light blue; the predicted mature peptides, AjKiss1a and AjKiss1b, are noted by the blue and green underlines. (B) The organization of the AjpreKiss gene is compared with the zebrafish and human preKiss genes. The exon-intron data were obtained from the respective genomic sequences from NCBI (MRZV01001091.1, NC_007122.7, NC_007115.7 and NC_000001.11). DNA structure is shown with exons numbered in green bands. ATG represents the start methionine codon and TGA represents the stop codon. (C) Alignment of the predicted echinoderm kisspeptin core sequences and functionally characterized chordate kisspeptins. Sequences of Holothuria scabra, Holothuria glaberrima, Strongylocentrotus purpuratus, and Asterias rubens kisspeptins were predicted by Elphick’s lab (Semmens and Elphick, 2017; Suwansa-Ard et al., 2018). Vertebrate kisspeptin core sequences were obtained from GenBank with detailed sequences listed in Figure 1—source data 1. The color align property was generated using Sequence Manipulation Suite online. The percentage of sequences that must agree for identity or similarity coloring was set as 40%. (D) Phylogenetic tree of the kisspeptin precursor and four different neuropeptide outgroups (Mirabeau and Joly, 2013). The tree was constructed on the basis of approximately Maximum-Likelihood algorithms using FastTree two with pre-trimmed sequences. Local support values (%) were provided using the Shimodaira-Hasegawa (SH) test and are indicated by numbers at the nodes. The detailed complete sequences are listed in Figure 1—source data 2 and trimmed sequences are listed in Figure 1—source data 3.
-
Figure 1—source data 1
Core sequences of kisspeptin from multiple species for alignment.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig1-data1-v1.txt
-
Figure 1—source data 2
Amino-acid sequences of the kisspeptin precursor and outgroups for phylogenetic analysis.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig1-data2-v1.txt
-
Figure 1—source data 3
Trimmed sequence alignment for phylogenetic tree construction.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig1-data3-v1.txt
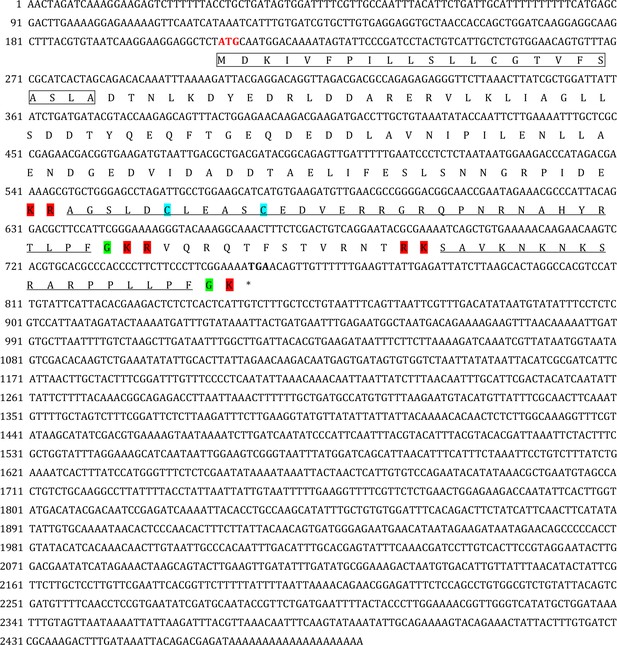
Gene structure of the Apostichopus japonicus kisspeptin precursor.
The signal peptide, predicted by the online SignalP-5.0 Server, is labeled in the box; the cleavage sites, predicted on the basis of previously known consensus cleavage motifs using the NeuroPred program, are highlighted in red; the glycine residues responsible for C-terminal amidation are highlighted in green; the cysteines paired in a disulfide-bonding structure are highlighted in light blue; and the predicted mature peptides with C-terminal amidation are underlined in black. The initiation codon (ATG) and the termination codon (TGA) are shown in bold.
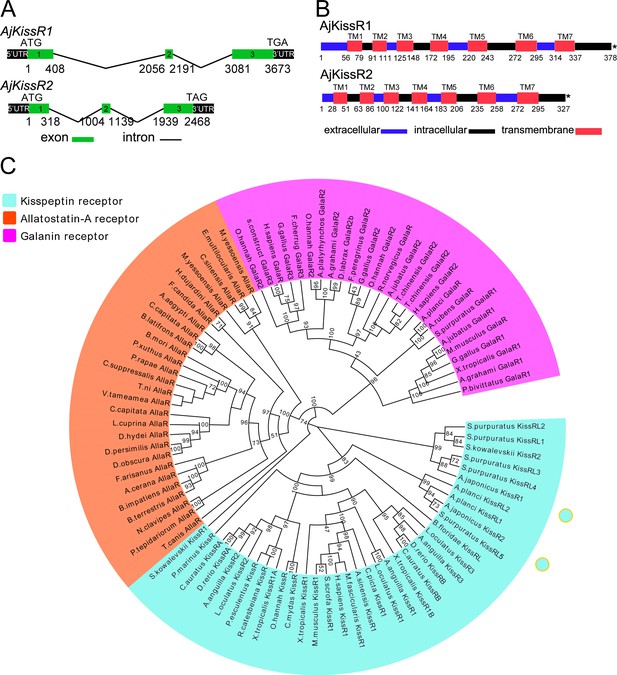
Gene structure and phylogenetic characterization of Apostichopus japonicus kisspeptin receptors (AjKissR1 and AjKissR2).
(A) DNA structures of AjKissR1 and AjKissR2. AjKissR1/2 DNA structure is shown with exons numbered in green bands. ATG represents the start methionine codon and TGA/TAG represents the stop codon. (B) Organization of the predicted protein structures. The seven transmembrane domains (TM1–TM7) are marked with red boxes. The N-terminal region and three extracellular (EC) rings are noted with blue boxes, and the C-terminal part and three intracellular (IC) rings are indicated with black boxes. Stop codons are represented by an asterisk. Arabic numbers under the bands indicate the nucleotide or amino acid sites. (C) Phylogenetic tree for kisspeptin, allatostatin-A and galanin receptors (KissR, AllaR and GalaR). The tree was constructed on the basis of approximately Maximum-Likelihood algorithms by FastTree two using AllaRs and GalaRs as outgroups (Ukena et al., 2014). Local support values were provided using the Shimodaira-Hasegawa (SH) test. The detailed sequences are listed in Figure 2—source data 1.
-
Figure 2—source data 1
Amino-acid sequences of kisspeptin receptors and outgroups for phylogenetic analysis.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig2-data1-v1.txt
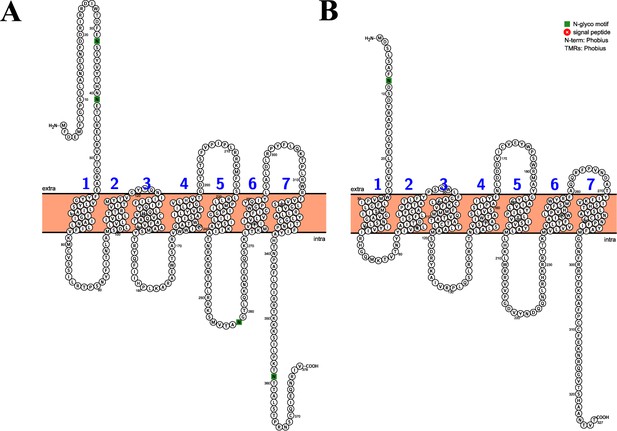
Sequence, topology and annotations of Apostichopus japonicus kisspeptin receptors (A: AjKissR1, B: AjKissR2) visualized using the Protter webservice.
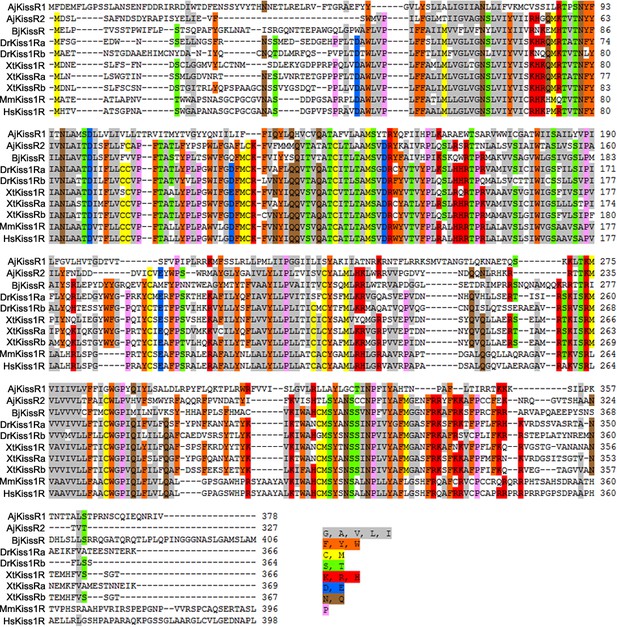
Alignment of the deduced Apostichopus japonicus kisspeptin receptor amino-acid sequences with functionally characterized chordate GPR54 molecules from other species.
Sequences of Branchiostoma japonicum kisspeptin receptor (BjKissR), Danio rerio kisspeptin receptors (DrKiss1Ra NP_001099149.2 and DrKiss1Rb NP_001104001.1), Xenopus tropicalis kisspeptin receptors (XtKiss1R NP_001163985.1, XtKissRa NP_001165296.1 and XtKissRb NP_001165295.1), Mus musculus kisspeptin receptor (MmKiss1R NP_444474.1), and Homo sapiens kisspeptin receptor (HsKiss1R NP_115940.2) were obtained from GenBank. Alignment was conducted using CLUSTAL W and the color align property was generated using Sequence Manipulation Suite online. The percentage of sequences that must agree for identity or similarity coloring was set as 60%.
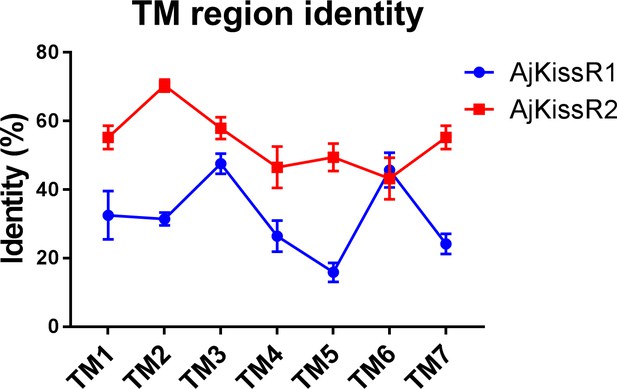
Transmembrane region sequence similarity of Apostichopus japonicus kisspeptin receptors to vertebrate kisspeptin receptors.
Figure 2—figure supplement 3—source data 1 lists the detailed identities.
-
Figure 2—figure supplement 3—source data 1
Primary metadata of detailed identities for Figure 2—figure supplement 3.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig2-figsupp3-data1-v1.xlsx
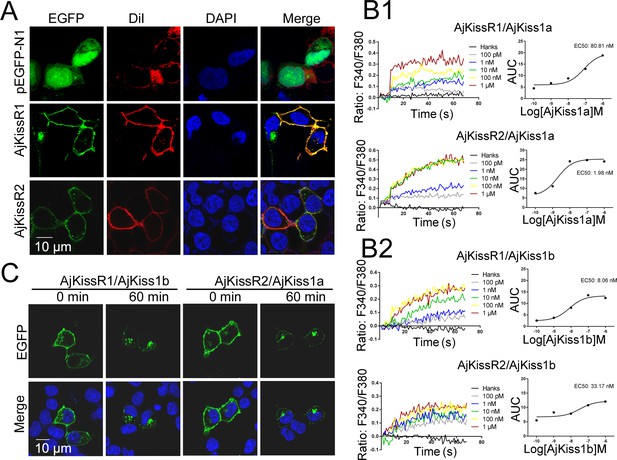
Functional characteristics of Apostichopus japonicus kisspeptins and receptors.
(A) The cells transiently expressing AjKissR1-EGFP or AjKissR2-EGFP were stained with cell membrane probe (DiI) and cell nucleus probe (DAPI) and detected by confocal microscopy. (B) After loading with Fura-2/AM, HEK293 cells expressing either FLAG-AjKissR1 or FLAG-AjKissR2 were exposed to the indicated concentrations of AjKiss1a (B1) and AjKiss1b (B2), and continuous fluorescence was recorded. AUC, Area Under the Curve. Figure 3B—source data 1 shows the primary metadata. (C) Internalization of AjKissR1-EGFP or AjKissR2-EGFP initiated by 1.0 μM of the indicated ligand was determined after a 60 min incubation by confocal microscopy. All pictures and data are representative of at least three independent experiments.
-
Figure 3—source data 1
Primary metadata of Ca2+ mobilization assay for Figure 3B.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig3-data1-v1.xlsx
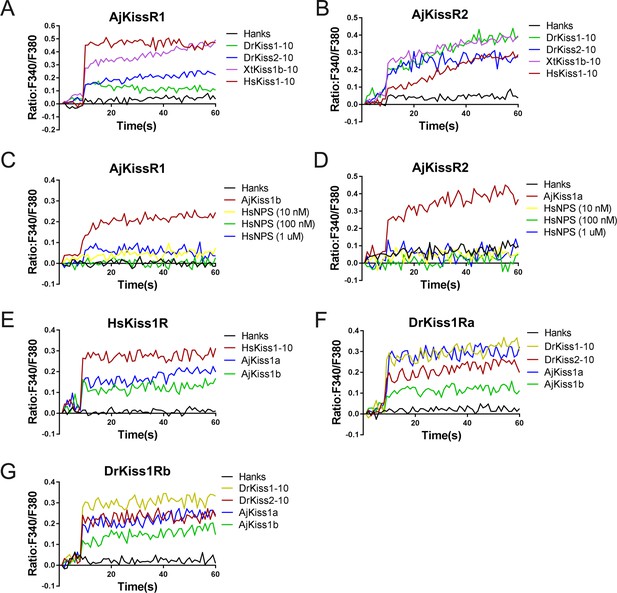
Functional cross-activity between the A. japonicus and vertebrate Kisspeptin/Kisspeptin receptor systems.
Intracellular Ca2+ mobilization in AjKissR1- (A) or AjKissR2-expressing (B) HEK293 cells was measured in response to 1.0 μM DrKiss1-10, DrKiss2-10, XtKiss1b-10, or HsKiss1-10 using Fura-2/AM. No Ca2+-mobilization-mediated activity was detected in AjKissR1- (C) or AjKissR2-expressing (D) HEK293 cells upon administration of the indicated concentrations of human neuropeptide S (HsNPS). Intracellular Ca2+ mobilization in human kisspeptin receptor (HsKiss1R)-expressing HEK293 cells was measured in response to 1.0 μM HsKiss1-10, AjKiss1a or AjKiss1b (E), as well as in zebrafish kisspeptin receptor (DrKiss1Ra or DrKiss1Rb)-expressing cells responding to 1.0 μM DrKiss1-10, AjKiss1a, or AjKiss1b (F, G). Figure 4—source data 1 shows the primary metadata. All data shown are representative of at least three independent experiments.
-
Figure 4—source data 1
Primary metadata of Ca2+ mobilization assay for Figure 4A-G.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig4-data1-v1.xlsx
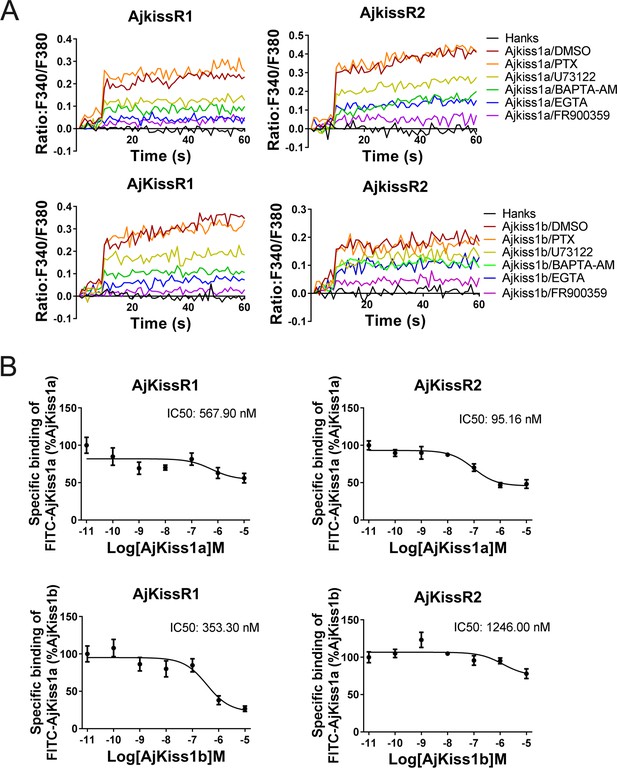
Apostichopus japonicus kisspeptin receptors are directly activated by kisspeptins via a Gαq-dependent pathway.
(A) Intracellular Ca2+ mobilization in AjKissR1- and AjKissR2-expressing HEK293 cells was measured in response to 100 nM AjKiss1a or AjKiss1b, using cells that had been pre-treated for 12 hr with Gαi protein inhibitor (PTX, 100 ng/mL), or for 1 hr with DMSO, Gαq protein inhibitor (FR900359, 1.0 μM), PLC inhibitor (U73122, 10 μM), intracellular calcium chelator (BAPTA-AM, 100.0 μM), or extracellular calcium chelator (EGTA, 5.0 mM). Figure 5—source data 1 presents the primary metadata. Pictures shown are representative of at least three independent experiments. (B) Competitive binding of 1.0 μM fluorescein isothiocyanate (FITC)-AjKiss1a to AjKissR1 or AjKissR2 in the presence of the indicated concentration of AjKiss1a or AjKiss1b. Error bars represent the SEM for at least three independent experiments.
-
Figure 5—source data 1
Primary metadata of Ca2+ mobilization assay and binding assay for Figure 5A and B.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig5-data1-v1.xlsx
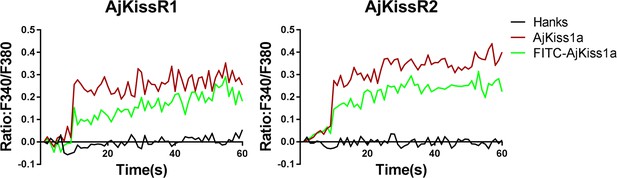
Functional activity of FITC-AjKiss1a evaluated by intracellular Ca2+ mobilization detection.
Intracellular Ca2+ mobilization in AjKissR1/2-expressing HEK293 cells was measured in response to 1.0 μM stimuli using Fura-2/AM.
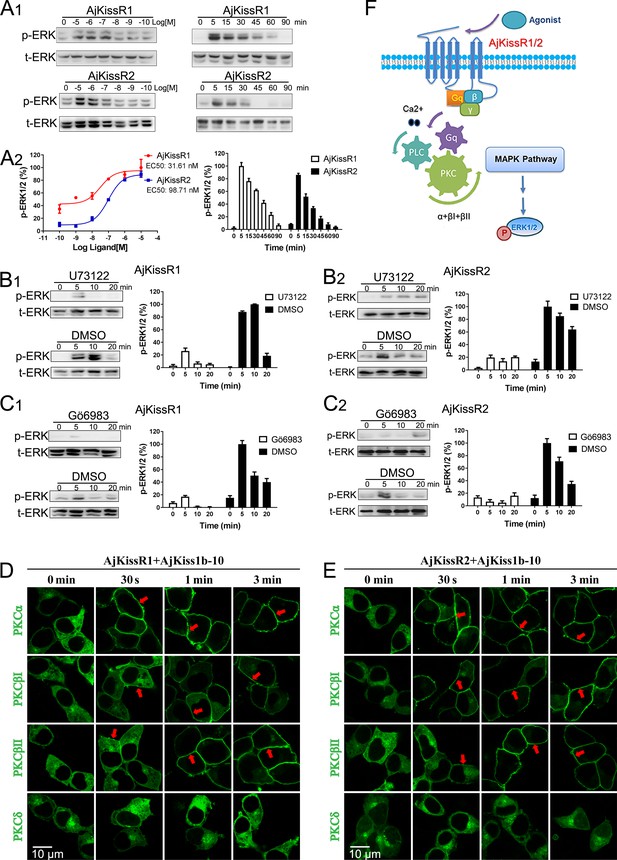
ERK1/2 activation mediated by AjKissR1 or AjKissR2.
(A) Concentration- and time-dependent effects of AjKiss1b-10 on ERK1/2 phosphorylation in HEK293 cells that were stably expressing FLAG–AjKissR1 or FLAG–AjKissR2. Cells were challenged with different concentrations of AjKiss1b-10 for 5 min or incubated with 1.0 μM AjKiss1b-10 for the indicated times. Immunoblots were quantified using a Bio-Rad Quantity One Imaging system. (B, C) ERK1/2 phosphorylation, activated by AjKiss1b-10, was blocked by PLC or PKC inhibitors. Serum-starved HEK293 cells expressing FLAG–AjKissR1 or FLAG–AjKissR2 were pre-treated with DMSO, PLC inhibitor (U73122, 10 μM), or PKC inhibitor (Gö6983, 10 μM) for 1 hr prior to AjKiss1b-10 stimulation (1.0 μM). (D, E) Role of various PKC isoforms in the activated signaling pathways of the sea cucumber kisspeptin receptor. HEK293 cells, co-transfected with FLAG–AjKissR1 or FLAG–AjKissR2 and different PKC-EGFP isoforms, were stimulated by 1.0 μM AjKiss1b-10 for the indicated times and then examined by confocal microscopy. Red arrows denote the recruitment of PKC–EGFP isoforms on the cell membrane. (F) Schematic diagram of agonist-induced A. japonicus kisspeptin receptor activation. AjKiss1b-10 binding to AjKissR1 or AjKissR2 activates the Gαq family of heterotrimeric G proteins, which leads to dissociation of the G protein subunits Gβγ, and activates PLC, leading to intracellular Ca2+ mobilization. This, in turn, activates PKC (isoform α and β) and stimulates the phosphorylation of ERK1/2. The ratio of p-ERK1/2 to total ERK1/2 was normalized to the peak value detected in the corresponding experiments (for example, the peak value of the ratio of AjKissR1/AjKiss1b-10 (10 μM) for the dose-dependent analysis, or of AjKissR1/AjKiss1b-10 (1.0 Aj) at 5 min for the time-course analysis). All pictures and data are representative of at least three independent experiments.
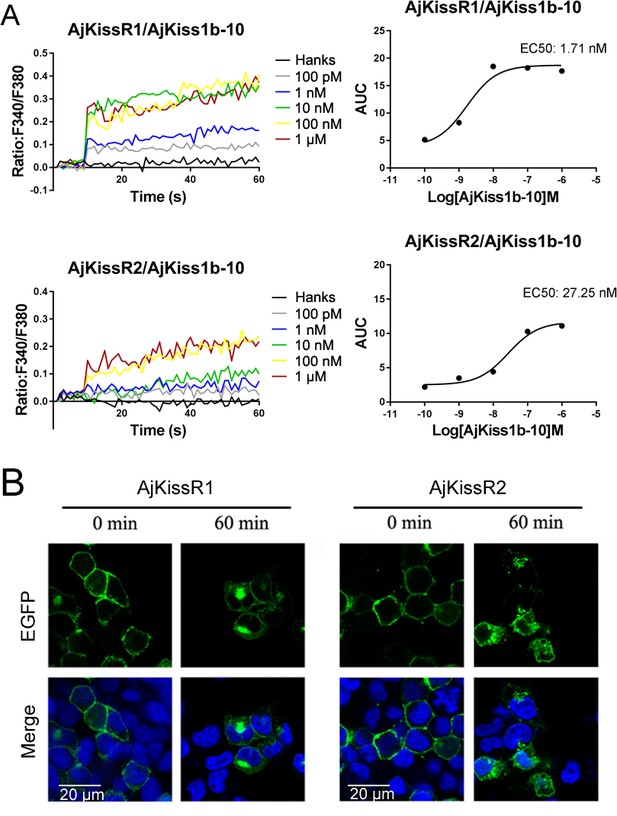
Functional activity of AjKiss1b-10.
(A) Intracellular Ca2+ mobilization in AjKissR1/2-expressing HEK293 cells was measured in response to AjKiss1b-10 at the indicated concentrations using Fura-2/AM. (B) Internalization of overexpressed AjKissR1/2 initiated by 1.0 μM AjKiss1b-10 in AjKissR1–EGFP- or AjKissR2–EGFP-expressing HEK293 cells was determined by confocal microscopy. All data shown are representative of at least three independent experiments.
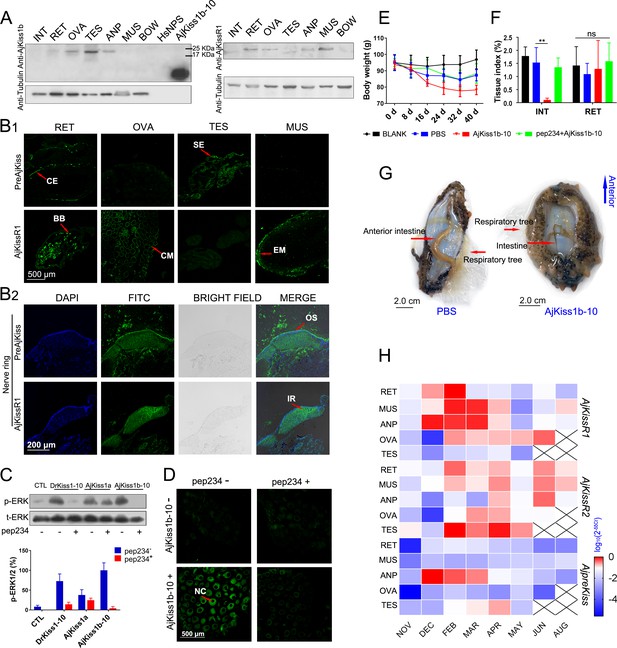
Physiological function analysis of kisspeptin signaling systems in Apostichopus japonicus.
(A) Western Blot analysis of A. japonicus kisspeptin precursor and kisspeptin receptor (AjKissR1) in different tissues of sea cucumber. INT, intestine; RET, respiratory tree; ANP, anterior part; OVA, ovary; TES, testis; MUS, muscle; and BOW, body wall. (B) Immunofluorescence histochemical staining of A. japonicus kisspeptin precursor and AjKissR1 in RET, OVA, TES, MUS (B1) and nerve ring (B2) of the sea cucumber. CE, coelomic epithelium; BB, brown body; CM, cell membrane; SE, spermatogenic epithelium; EM, epithelium of muscle; OS, outer surface; and IR, internal region. (C) ERK1/2 phosphorylation activity of kisspeptins and the inhibitory effect of a vertebrate kisspeptin antagonist (pep234) on the cultured ovary of sea cucumber. Samples were collected and fixed after 2 hr of ligand administration with or without a 4-hr pre-treatment with pep234, in optimized L15 medium at 18°C. Error bars represent SEM for three independent experiments. Immunoblots were quantified using a Bio-Rad Quantity One Imaging system. (D) Immunofluorescence histochemical staining of p-ERK signal in cultured oocytes of sea cucumber. Samples were collected and fixed after 2 hr of ligand administration with or without a 4-hr pretreatment with pep234, in optimized L15 medium at 18°C. NC indicates the nucleus of oocytes. (E, F) Variation of body weight and tissue index (ratio of tissue weight/body weight) over 40 days of stimulus treatment. Each symbol and vertical bar represent mean ± SEM (n = 5 animals). ** indicates extremely significant differences (p=0.0001), as demonstrated by one-way ANOVA followed by Tukey’s multiple comparisons test. (G) Degenerated intestine in AjKiss1b-10 treated sea cucumbers. (H) Heatmap showing the expression profile of A. japonicus kisspeptin and kisspeptin receptors (AjKissR1/R2 and AjKiss1) in different tissues and developmental stages of sea cucumber. The variation in color represents the relative expression level of each gene in different samples (normalized against the peak values in all samples and logarithmized). The number of animals used for all samples is six, except for the number of ovary samples, with one in NOV (November) and JUN (June), three in DEC (December) and FEB (February), five in MAR (March), and six in APR (April) and MAY (May), and in testis, with two in NOV (November) and DEC (December), four in FEB (February) and MAR (March), and six in APR (April) and MAY (May). Figure 7—source data 1 represents the primary metadata. All pictures and data are representative of at least three independent experiments.
-
Figure 7—source data 1
Primary metadata of body weight, tissue index and qPCR assay for Figure 7E, F and H.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig7-data1-v1.xlsx
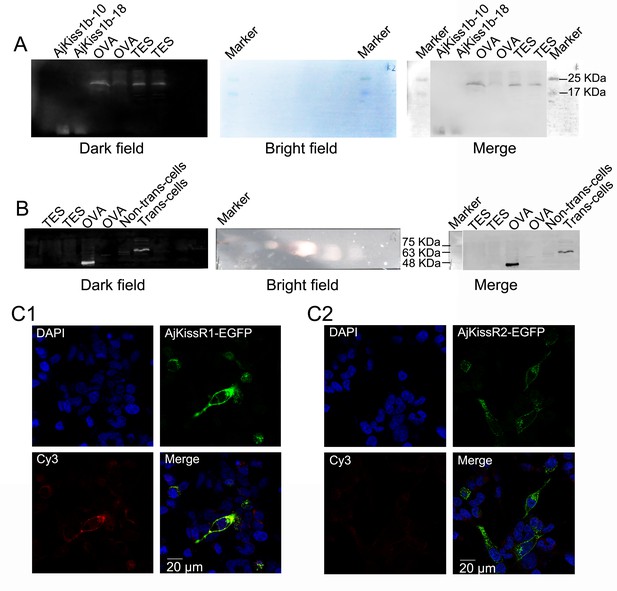
Antibody specificities of anti-AjKiss1b-10 and anti-AjKissR1 IgG antibodies.
(A) Western Blot analysis showing the specificity of the anti-AjKiss1b-10 IgG polyclonal antibody. AjKiss1b-10 and AjKiss1b (20 μL, 100 nM) were used as positive controls. (B) Antibody specificities of the anti-AjKissR1 IgG polyclonal antibody. AjKissR1–EGFP transfected or non-transfected cells were also tested. (C) Confocal microscopy analysis of the specificity of the anti-AjKissR1 IgG polyclonal antibody. AjKissR2–EGFP expressing cells were stained and scoped as a negative control. Cy3-conjugated goat anti-rabbit IgG was used as a secondary antibody. All data shown are representative of at least three independent experiments.

General morphology and histology of Apostichopus japonicus tissues.
(A–D) Light micrographs of H and E stained sections of respiratory tree, ovary, testis and muscle. CE, coelomic epithelium; BB, brown body; CM, cell membrane; NC, nucleus of oocytes; NU, nucleolus of oocytes; SE, spermatogenic epithelium; EM, epithelium of muscle; and MC, myocyte. (E) Gross anatomy of the anterior part (ANP). (F) Light micrographs of H and E stained sections of ANP and histology of nerve ring (NR). OS, outer surface; IR, internal region; CR, calcareous ring; AX, axon of neuron; and CB, cell body of neuron. All data shown are representative of at least three independent experiments.
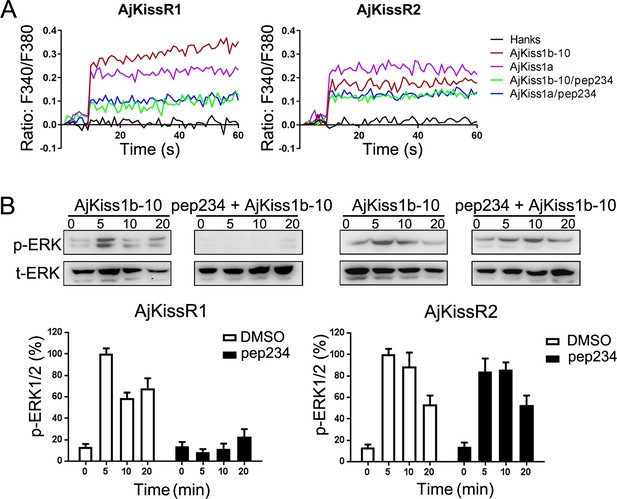
Inhibitory effect of pep234 on AjKissR1 and AjKissR2 activation.
(A) Intracellular Ca2+ mobilization in AjKissR1- and AjKissR2-expressing HEK293 cells was measured in response to 100 nM AjKiss1a or AjKiss1b-10 pre-treated with DMSO or the KISS1 antagonist pep234 (1.0 μM). (B) ERK1/2 phosphorylation activity of kisspeptins and the inhibitory effect of pep234 in AjKissR1- and AjKissR2-expressing HEK293 cells. Samples were measured after 2 hr of ligand administration with or without pep234 pre-treatment. Error bars represent SEM for three independent experiments. Immunoblots were quantified using a Bio-Rad Quantity One Imaging system. All data shown are representative of at least three independent experiments.
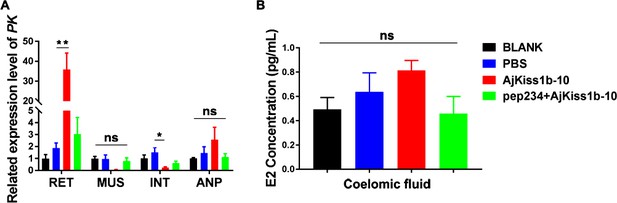
Functional activity of AjKiss1b-10 in Apostichopus japonicus.
(A) Expressional change of the gene encoding the glycolytic enzyme pyruvate kinase (PK) in tissues of sea cucumbers responds to a 40-day administration of AjKiss1b-10. RET, respiratory tree; MUS, muscle; INT, intestine; and ANP, anterior part of sea cucumber. (B) E2 concentration in the coelomic fluid of sea cucumbers did not significantly respond to AjKiss1b-10. Each error bar represents SEM (n = 5 biological replicates). * indicates significant difference (PBS vs. AjKiss1b-10, p=0.0188), and ** indicates extremely significant difference (PBS vs. AjKiss1b-10, p=0.0002), as determined by ANOVA followed by Tukey’s multiple comparisons test. Figure 7—figure supplement 4—source data 1 lists the detailed primary metadata and statistics.
-
Figure 7—figure supplement 4—source data 1
Primary metadata of qPCR assay and E2 concentration for Figure 7—figure supplement 4A and B.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig7-figsupp4-data1-v1.xlsx

Mean body weight (A) and tissue index (B) change over annual investigation.
Relative gut mass (RGM) and relative ovary weight (ROW) indicate the ratio of tissue weight/body weight. The number of animals used for all samples is six, except for the number of ovary samples, with one in NOV (November), three in DEC (December), FEB (February) and JUN (June), five in MAR (March), and six in APR (April) and MAY (May). Figure 7—figure supplement 5—source data 1 lists the primary metadata.
-
Figure 7—figure supplement 5—source data 1
Primary metadata of body weight and tissue index in annual investigation for Figure 7—figure supplement 5A and B.
- https://cdn.elifesciences.org/articles/53370/elife-53370-fig7-figsupp5-data1-v1.xlsx
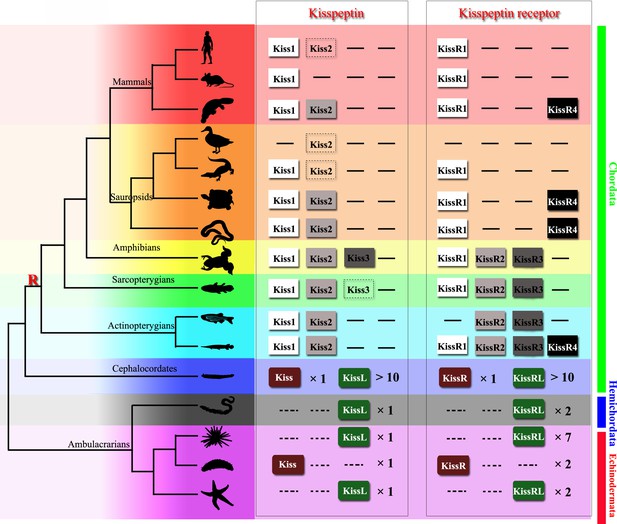
Recently identified Kisspeptin or Kisspeptin receptor genes among some deuterostomes.
The species, indicated by silhouette images downloaded from the PhyloPic database, were clustered in a phylogenetic tree and classified by different colors. Red highlighted ‘R’ indicates a whole-genome duplication event. Kiss/KissR indicates the identified Kisspeptin/Kisspeptin receptor gene, and KissL/KissRL indicates a predicted Kisspeptin-like/Kisspeptin-like receptor gene. Dashed symbols indicate pseudogenes. Arabic numerals indicate the number of genes identified or predicted from public data. The evolutionary tree of the indicated species was modified from Pasquier et al., 2014. Image credits: all silhouettes from PhyloPic, human by T Michael Keeseyacorn; mouse by Anthony Caravaggi; platypus by Sarah Werning; duck by Sharon Wegner-Larsen; crocodile by B Kimmel; turtle by Roberto Díaz Sibaja; python by V Deepak; frog uncredited; coelacanth by Yan Wong; zebrafish by Jake Warner; spotted gar by Milton Tan; Branchiostoma by Mali'o Kodis, photograph by Hans Hillewaert; acorn worm by Mali'o Kodis, drawing by Manvir Singh; starfish by Hans Hillewaert and T Michael Keesey; sea cucumber by Lauren Sumner-Rooney; sea urchin by Jake Warner.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293 cell line | The National Institutes of Health (Bethesda, MD) | RRID:CVCL_0045 | Cell line maintained in this lab. |
| Antibody | Anti-phospho-ERK1/2(Thr202/Tyr204) (monoclonal, rabbit) | Cell Signaling Technology | CAT#9101 RRID:AB_331646 | IF (1:2000) |
| Antibody | Anti-ERK1/2 antibody (monoclonal, rabbit) | Cell Signaling Technology | CAT#9102 RRID:AB_330744 | IF (1:2000) |
| Antibody | Anti-beta Tubulin (monoclonal, rabbit) | Beyotime | CAT#AF1216 | IF (1:2000) |
| Antibody | FITC-conjugated goat anti-rabbit IgG (polyclonal, goat) | Beyotime | CAT#A0562 | IF (1:500) |
| Antibody | Cy3-conjugated goat anti-rabbit IgG (polyclonal, goat) | Beyotime | CAT#A0516 | IF (1:500) |
| Antibody | HRP-conjugated goat anti-rabbit IgG (polyclonal, goat) | Beyotime | CAT#A0208 | IF (1:500) |
| Antibody | anti-AjKiss1b-10 IgG (polyclonal, rabbit) | ChinaPeptides | CNE180821096 | Antigen sequence: CSRARPPLLPF-NH2 IF (1:1000) |
| Antibody | anti-AjKissR1 Ser150~Trp174IgG (polyclonal, rabbit) | Wuhan Transduction Bio | PC059 | Antigen sequence: SYTRYQFIIHPLKARAEWTSARVWW IF (1:1000) |
| Sequence-based reagent | Primers for plasmid construction AjKissR1–EGFP | This paper | PCR PRIMER | FORWARD CGAATTCATGTTTGACGAAATGTTC EcoR I |
| Sequence-based reagent | Primers for plasmid construction AjKissR1–EGFP | This paper | PCR PRIMER | REVERSE GTGGATCCCGAACGATACGATTCTGTTC BamH I |
| Sequence-based reagent | Primers for plasmid construction FLAG–AjKissR1 | This paper | PCR PRIMER | FORWARD GGAATTCATGTTTGACGAAATGTTC EcoR I |
| Sequence-based reagent | Primers for plasmid construction FLAG–AjKissR1 | This paper | PCR PRIMER | REVERSE CGGGATCCTCAAACGATACGATTCTGTTC BamH I |
| Sequence-based reagent | Primers for plasmid construction AjKissR2–EGFP | This paper | PCR PRIMER | FORWARD CGAATTCATGGACAGCCTCTCAGC EcoR I |
| Sequence-based reagent | Primers for plasmid construction AjKissR2–EGFP | This paper | PCR PRIMER | REVERSE CCGTCGACTGAGTTACAGTATTTGCTG SalI |
| Sequence-based reagent | Primers for plasmid construction FLAG–AjKissR2 | This paper | PCR PRIMER | FORWARD CCAAGCTTGGATGGACAGCCTCTCAGCGTT Hind III |
| Sequence-based reagent | Primers for plasmid construction FLAG–AjKissR2 | This paper | PCR PRIMER | REVERSE CGGGATCCCGTGAGTTACAGTATTTGCTGCAT Bam HI |
| Sequence-based reagent | Primers for plasmid construction FLAG–HsKiss1R | This paper | PCR PRIMER | FORWARD CCAAGCTTGGATGCACACCGTGGCTAC Hind III |
| Sequence-based reagent | Primers for plasmid construction FLAG–HsKiss1R | This paper | PCR PRIMER | REVERSE CGGGATCCTCAGAGAGGGGCGTTGTCCT Bam HI |
| Sequence-based reagent | Primers for qPCR assays AjKissR1 | This paper | PCR PRIMER | FORWARDAGTGGACATCTGCAAGAGTATGG |
| Sequence-based reagent | Primers for qPCR assays AjKissR1 | This paper | PCR PRIMER | REVERSE CTTCCTGCGTAATGGTATCGGTA |
| Sequence-based reagent | Primers for qPCR assays AjKissR2 | This paper | PCR PRIMER | FORWARD TCTCGTTGTTGTCTTGACGTTTG |
| Sequence-based reagent | Primers for qPCR assays AjKissR2 | This paper | PCR PRIMER | REVERSETCGTCTGAAGTTTTCTCCCATGA |
| Sequence-based reagent | Primers for qPCR assays AjpreKiss | This paper | PCR PRIMER | FORWARD CCTACTGTCATTGCTCTGTGGAAC |
| Sequence-based reagent | Primers for qPCR assays AjpreKiss | This paper | PCR PRIMER | REVERSECAAGGTCATCTTCGTCTTGTTCTC |
| Sequence-based reagent | Primers for qPCR assays β-tubulin | This paper | PCR PRIMER | FORWARD CACCACGTGGACTCAAAATG |
| Sequence-based reagent | Primers for qPCR assays β-tubulin | This paper | PCR PRIMER | REVERSE GAAAGCCTTACGACGGAACA |
| Sequence-based reagent | Primers for qPCR assays β-actin | This paper | PCR PRIMER | FORWARD AAGGTTATGCTCTTCCTCACGC |
| Sequence-based reagent | Primers for qPCR assays β-actin | This paper | PCR PRIMER | REVERSE GATGTCACGGACGATTTCACG |
| Recombinant DNA reagent | AjKissR1–EGFP | This paper | Plasmid | C-terminal EGFP-tag, backbone pEGFP-N1 |
| Recombinant DNA reagent | FLAG-–AjKissR1 | This paper | Plasmid | N-terminal FLAG-tag, backbone pCMV–FLAG |
| Recombinant DNA reagent | AjKissR2–EGFP | This paper | Plasmid | C-terminal EGFP-tag, backbone pEGFP-N1 |
| Recombinant DNA reagent | FLAG–AjKissR2 | This paper | Plasmid | N-terminal FLAG-tag, backbone pCMV–FLAG |
| Recombinant DNA reagent | FLAG–HsKiss1R | This paper | Plasmid | N-terminal FLAG-tag, backbone pCMV–FLAG |
| Recombinant DNA reagent | FLAG–DrKiss1Ra | This paper | Plasmid | N-terminal FLAG-tag, backbone pCMV–FLAG |
| Recombinant DNA reagent | FLAG–DrKiss1Rb | This paper | Plasmid | N-terminal FLAG-tag, backbone pCMV–FLAG |
| Recombinant DNA reagent | EGFP-tagged rat PKC isoforms (α, βI, βII and δ) | Kindly provided byDr Jin O-Uchi, University of Rochester and Dr Naoaki Sato, Kobe University | Plasmid | C-terminal EGFP-tag, backbone pTB701 |
| Peptide, recombinant protein | AjKiss1a (C-terminal amidated) | This paper | AGSLDc < CLEASC > EDVERRGRQPNRNAHYRTLPF-NH2 | |
| Peptide, recombinant protein | FITC–AjKiss1a (C-terminal amidated) | This paper | FITC–AGSLDc < CLEASC > EDVERRGRQPNRNAHYRTLPF-NH2 | |
| Peptide, recombinant protein | AjKiss1a-15 (C-terminal amidated) | This paper | GRQPNRNAHYRTLPF-NH2 | |
| Peptide, recombinant protein | AjKiss1a-10 (C-terminal amidated) | This paper | AHYRTLPF-NH2 | |
| Peptide, recombinant protein | AjKiss1b (C-terminal amidated) | This paper | SAVKNKNKSRARPPLLPF-NH2 | |
| Peptide, recombinant protein | AjKiss1b-10 (C-terminal amidated) | This paper | SRARPPLLPF-NH2 | |
| Peptide, recombinant protein | HsKISS1-10 (C-terminal amidated) | This paper | YNWNSFGLRF-NH2 | |
| Peptide, recombinant protein | XtKISS3/KISS1b-10 (C-terminal amidated) | This paper | YNVNSFGLRF-NH2 | |
| Peptide, recombinant protein | DrKISS1-10 (C-terminal amidated) | This paper | YNLNSFGLRY-NH2 | |
| Peptide, recombinant protein | DrKISS2-10 (C-terminal amidated) | This paper | FNYNPFGLRF-NH2 | |
| Peptide, recombinant protein | pep234 (C-terminal amidated) | This paper | ac-(D-A)NWNGFG(D-W)RF-NH2 | |
| Commercial assay or kit | Rapid DNA Ligation kit | Beyotime | CAT#D7003 | |
| Commercial assay or kit | SYBR PrimeScript RT reagent Kit | TaKaRa | CAT #RR037A | |
| Commercial assay or kit | Iodine (125I) radioimmunoassay kit | Beijing North Institute of Biotechnology | S10940094 | |
| Chemical compound, drug | Pertussis toxin (PTX) | Tocris Bioscience | Cat #3097/50U | Specific inhibitor of Gαi |
| Chemical compound, drug | U73122 | Tocris Bioscience | Cat #1268/10 | PLC inhibitor |
| Chemical compound, drug | BAPTA-AM | Tocris Bioscience | Cat #2787/25 | Intracellular calcium chelator |
| Chemical compound, drug | EGTA | Tocris Bioscience | Cat #2807/1G | Extracellular calcium chelator |
| Chemical compound, drug | Gö6983 | Tocris Bioscience | Cat #2285/1 | Broad spectrum PKC inhibitor |
| Chemical compound, drug | FR900359 | Kindly provided by Dr Shihua Wu, Zhejiang University | Specific inhibitor of Gαq |





