Virus infection is controlled by hematopoietic and stromal cell sensing of murine cytomegalovirus through STING
Figures
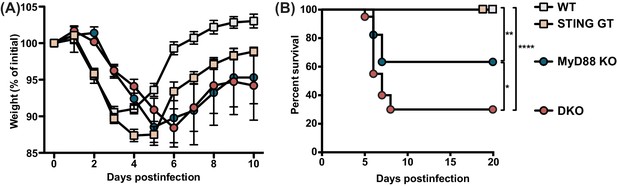
MyD88 and STING control morbidity and mortality during MCMV infection.
Mice were infected with 50,000 PFU MCMV WT-1, weight loss and survival was monitored over time. (A) Weight loss over time in wildtype (n = 12), STING-deficient (STING GT, n = 21), MyD88-deficient (MyD88 KO, n = 9) and mice deficient in both STING and MyD88 (DKO; n = 14). The numbers indicate the number of mice at the start of the experiment, weight loss of surviving mice at each timepoint is plotted. (B) Survival curves of wildtype (n = 17), STING GT (n = 18), MyD88 KO (n = 17) and DKO mice (n = 20). Cumulative data of 3 independent experiments. Error bars indicate SEM; *p<0.05, **p<0.01, ****p<0.0001.
-
Figure 1—source data 1
MyD88 and STING control morbidity and mortality during MCMV infection.
- https://cdn.elifesciences.org/articles/56882/elife-56882-fig1-data1-v2.xlsx
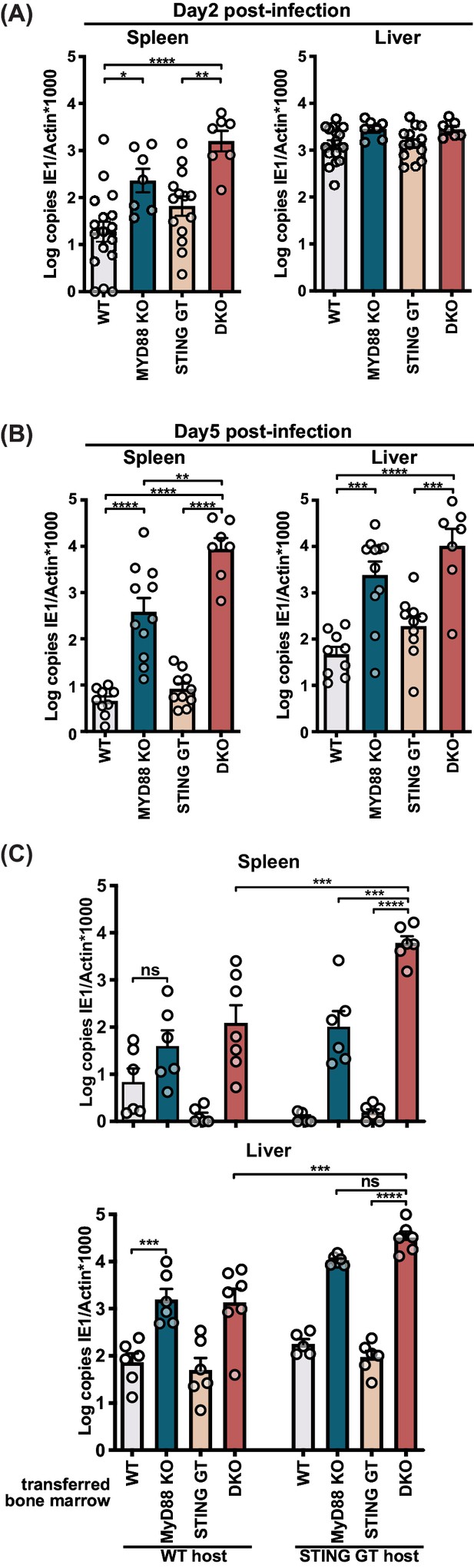
STING contributes to control of MCMV in the hematological and stromal compartment, whereas MyD88 in the hematological compartment potently controls infection.
Mice were infected with 50,000 PFU (A) and (B) or 20,000 PFU (C) MCMV. Viral load was quantified 2 days (A) or 5 days (B) and (C) p.i. (C) Indicated bone marrow was adoptively transferred into irradiated wildtype (WT) or STING-deficient (STING GT) hosts. Bone marrow chimeras were infected 6 weeks post transfer and viral load was analyzed 5 days p.i. Each panel shows cumulative data of 2 independent experiments. Error bars indicate SEM; ns, not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
-
Figure 2—source data 1
STING contributes to control of MCMV in the hematological and stromal compartment, whereas MyD88 in the hematological compartment potently controls infection.
- https://cdn.elifesciences.org/articles/56882/elife-56882-fig2-data1-v2.xlsx
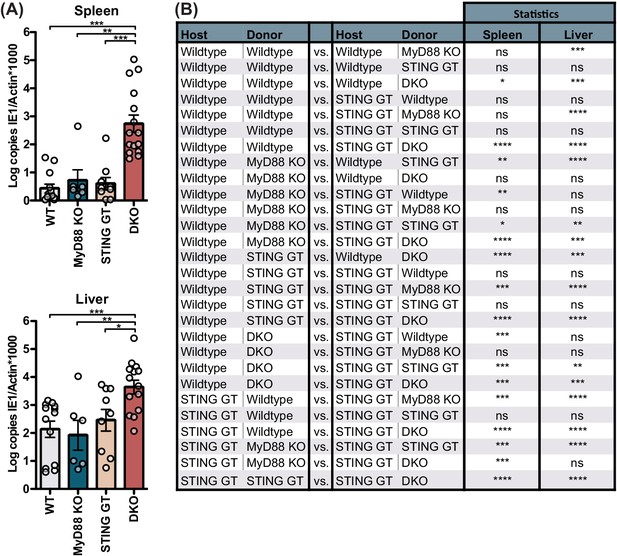
Viral load for 20,000 PFU infection 5 days p.i. and extended statistical analysis for bone marrow chimeras.
(A) Mice were infected with 20,000 PFU and viral load was quantified 5 p.i. (B) Extended statistical analysis for bone marrow chimeras presented in Figure 2C. Each panel shows cumulative data of 2 independent experiments. Error bars indicate SEM; ns, not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
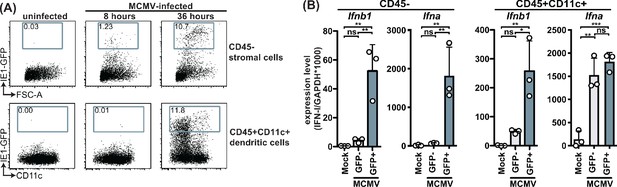
MCMV-infected cells specifically produce IFNβ upon infection.
WT mice were infected with 100,000 PFU MCMV IE1-GFP reporter virus. (A) Analysis of GFP expression in CD45- stromal cells and CD45+CD11c+ DC at 8 hr and 36 hours p.i. (B) GFP+ and GFP- stromal cells and DC were FACS-sorted 36 hours p.i. and Ifnb1 and pan-Ifna transcript levels were quantified by real-time PCR. Both panels show representative experiments from two independent experiments. Error bars indicate SD; ns, not significant, *p<0.05, **p<0.01, ***p<0.001.
-
Figure 3—source data 1
MCMV-infected cells specifically produce IFNβ upon infection.
- https://cdn.elifesciences.org/articles/56882/elife-56882-fig3-data1-v2.xlsx
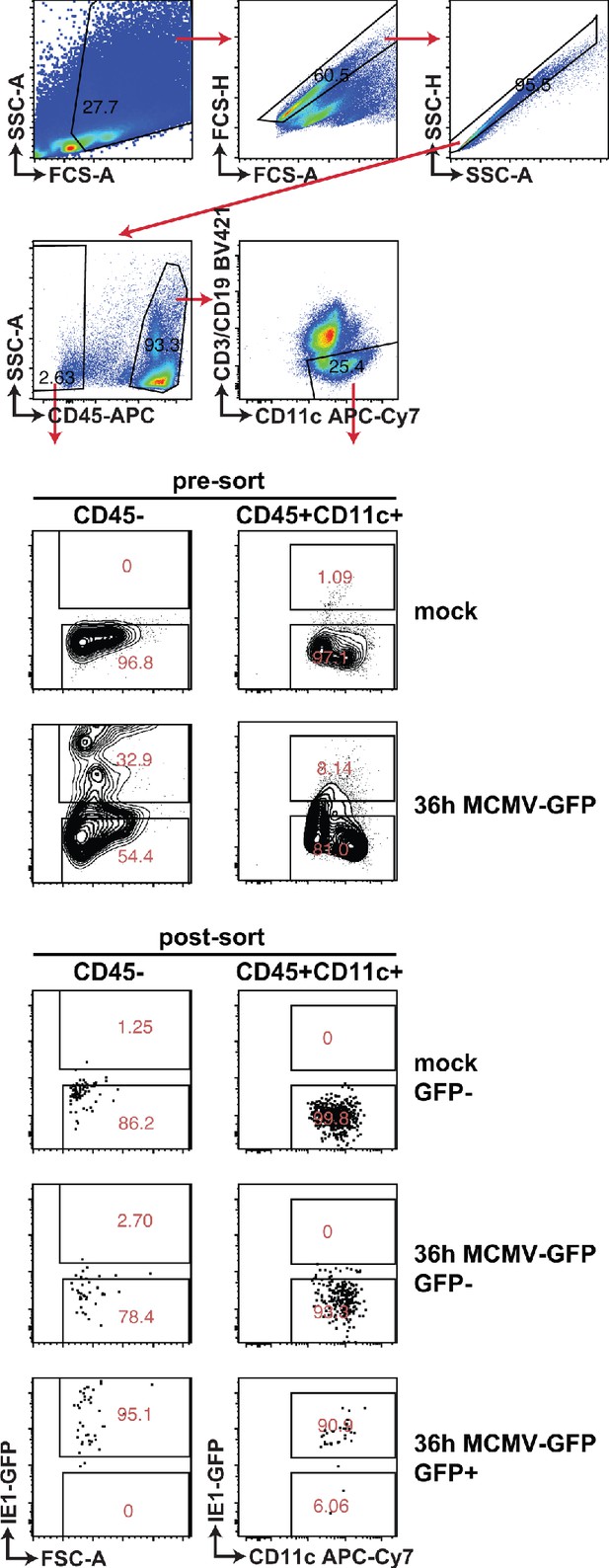
Gating strategy and purity of sorted cell populations.
WT mice were infected with 100,000 PFU MCMV IE1-GFP reporter virus. GFP+ and GFP- stromal cells and DC were FACS-sorted 36 hours p.i. Representative gating strategy and purity of sorted cells that were used in Figure 3BC are shown.
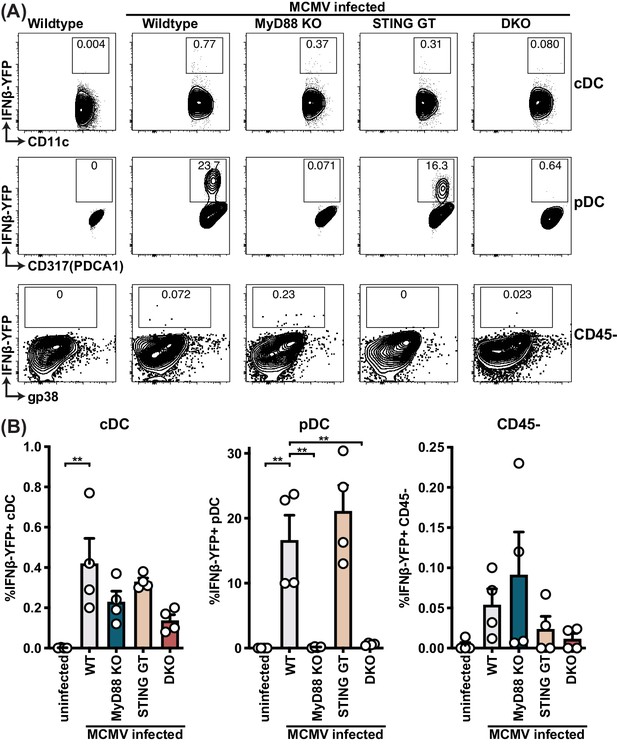
pDCs produce IFNβ in a MyD88-dependent but STING-independent manner in IFNβ-YFP reporter mice.
IFNβ-YFP reporter mice were backcrossed to MyD88- (MyD88 KO), STING- (STING GT) and double-deficient (DKO) mice. Animals were infected with 200,000 PFU WT1 MCMV and analyzed 48 hr post infection. Spleens were digested to a single cell suspension, stained and analyzed by flow cytometry. Error bars indicate SD; **p<0.01.
-
Figure 4—source data 1
pDCs produce IFNβ in a MyD88-dependent but STING-independent manner in IFNβ-YFP reporter mice.
- https://cdn.elifesciences.org/articles/56882/elife-56882-fig4-data1-v2.xlsx
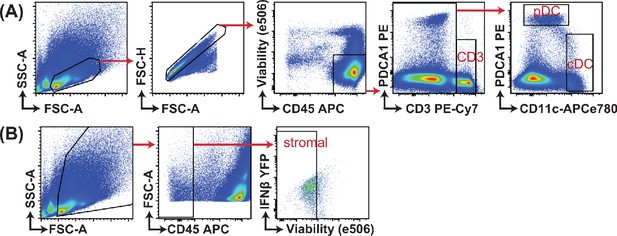
Gating strategy for analysis of IFNβ-YFP reporter mice.
IFNβ-YFP reporter mice were backcrossed to MyD88- (MyD88 KO), STING- (STING GT) and double-deficient (DKO) mice. Animals were infected with 200,000 PFU WT1 MCMV and analyzed 48 hr post infection. Gating strategy for samples presented in Figure 4 are shown.
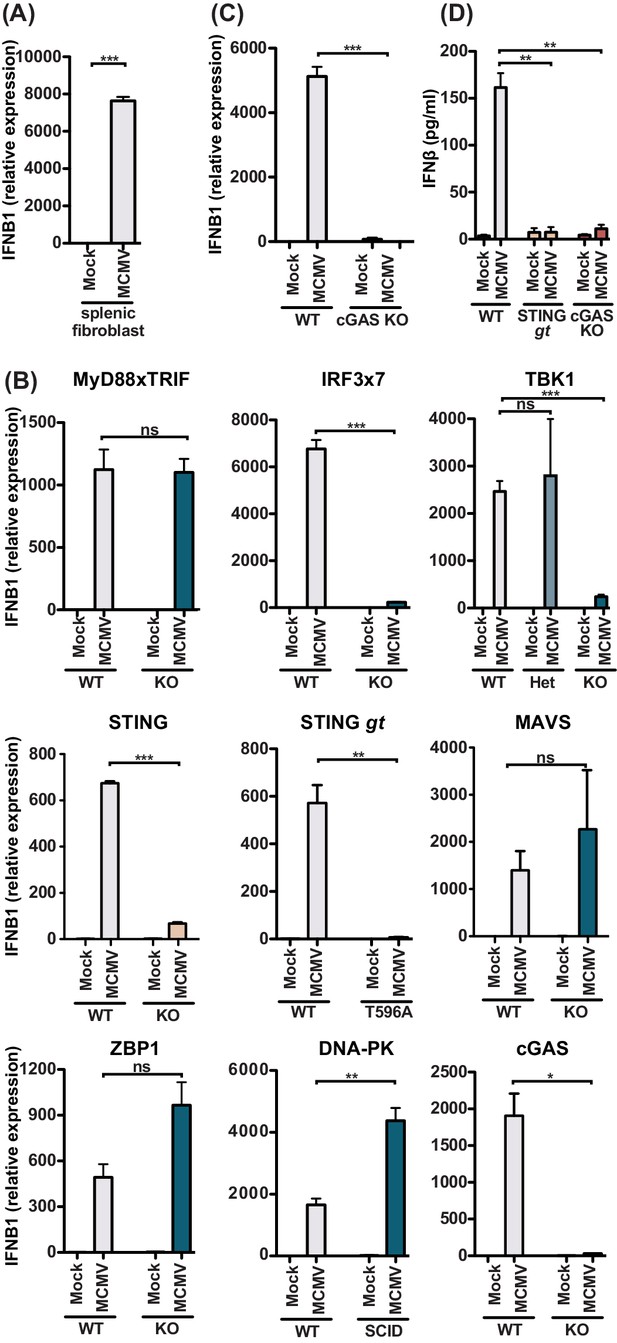
MCMV-induced fibroblast IFNβ is triggered by cGAS-STING-dependent but MyD88-Trif-MAVS-independent mechanisms.
(A) IFNB1 mRNA levels of primary splenic fibroblasts infected with WT1 MCMV (MOI = 5) 8 hr post-infection. (B) IFNB1 mRNA levels of murine embryonic fibroblasts (MEF) from wildtype (WT) or indicated deficient mice were infected and analyzed as in (A). (C) IFNB1 mRNA levels in infected WT or cGAS-deficient primary splenic fibroblasts, analyzed as in (A). (D) Secreted IFNβ by WT or indicated gene deficient MEF, infected with MCMV (MOI = 0.5); supernatant was analyzed 48 hours p.i. by ELISA. Panels show representative experiments from two independent experiments performed in duplicate. WT, STING GT, and TBK1-, MAVS-, ZBP1-, DNA-PK-, and cGAS-deficient MEF represent data from two independent MEF preparations. Error bars indicate SEM; ns, not significant, *p<0.05, **p<0.01, ***p<0.001.
-
Figure 5—source data 1
MCMV-induced fibroblast IFNβ is triggered by cGAS-STING-dependent but MyD88-Trif-MAVS-independent mechanisms.
- https://cdn.elifesciences.org/articles/56882/elife-56882-fig5-data1-v2.xlsx
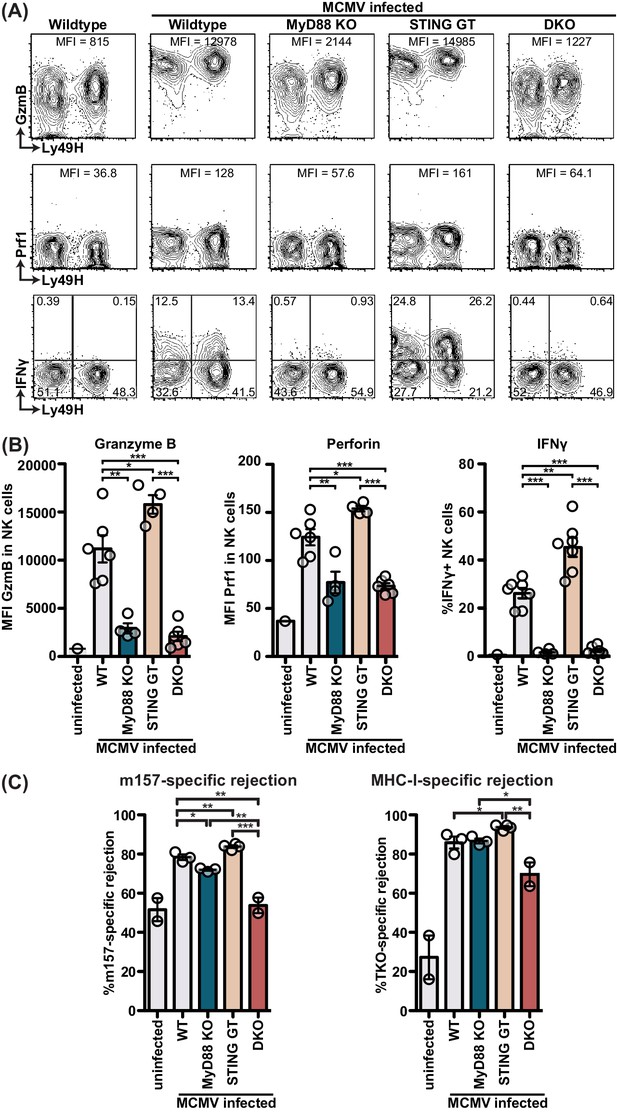
MyD88 and STING are required for NK cell cytolytic capacity during MCMV infection.
(A and B) Mice deficient in MyD88 and/or STING were infected with MCMV and 2 days later splenocytes were harvested and analyzed for GzmB, Prf1, and IFNγ expression by FACS. Representative contour plots of individual mice are shown in (A) and quantification for multiple mice is shown in (B). (C) Differentially labelled WT, m157-Tg and MHC-I deficient splenocytes were adoptively transferred into indicated day 3-infected mice. Specific rejection was analyzed 3 hr post-transfer in the spleen. Representative experiments from two independent experiments per panel are shown. Error bars indicate SEM; ns, not significant, *p<0.05, **p<0.01, ***p<0.001.
-
Figure 6—source data 1
MyD88 and STING are required for NK cell cytolytic capacity during MCMV infection.
- https://cdn.elifesciences.org/articles/56882/elife-56882-fig6-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, C57BL/6 background (Mus musculus) | C57BL/6 | Charles River Laboratories | 556; RRID:MGI:2160593 | |
| Strain, BALB/c background (Mus musculus) | BALB/c | Charles River Laboratories | 555; RRID:MGI:2160915 | |
| Strain, C57BL/6 background (Mus musculus) | STING golden ticket | Jackson Laboratories | 017537; RRID:IMSR_JAX:017537 | |
| Strain, C57BL/6 background (Mus musculus) | IFNβ-YFP reporter mice | Jackson Laboratories | 010818; RRID:IMSR_JAX:010818 | |
| Strain, C57BL/6 background (Mus musculus) | DNA-PK SCID | Jackson Laboratories | 001913; RRID:IMSR_JAX:001913 | |
| Strain, C57BL/6 background (Mus musculus) | Β2m KO | Jackson Laboratories | 002087; RRID:IMSR_JAX:002087 | |
| Strain, C57BL/6 background (Mus musculus) | M157-Tg | Tripathy et al., 2008 | ||
| Strain, C57BL/6 background (Mus musculus) | H-2Kb x H-2Db KO | Taconic | 4215; RRID:IMSR_TAC:4215 | |
| Strain, C57BL/6 background (Mus musculus) | MyD88 KO | S. Akira | RRID:MGI:3577712 | through the JCRB Laboratory Animal Resource Bank of the National Institute of Biomedical Innovation |
| Strain, C57BL/6 background (Mus musculus) | TBK1 KO | S. Akira | nbio156; RRID:MGI:3053427 | through the JCRB Laboratory Animal Resource Bank of the National Institute of Biomedical Innovation |
| Strain, C57BL/6 background (Mus musculus) | ZBP1 KO | S. Akira | nbio155; RRID:MGI:3776852 | through the JCRB Laboratory Animal Resource Bank of the National Institute of Biomedical Innovation |
| Strain, C57BL/6 background (Mus musculus) | IPS1 KO | Michael Gale | ||
| Strain, C57BL/6 background (Mus musculus) | cGAS KO | Herbert Virgin | ||
| Other | IRF3/7 KO MEF | Michael Diamond | Primary murine embryonic fibroblasts. | |
| Other | STING KO MEF | Glen Barber | Primary murine embryonic fibroblasts. | |
| Other | MyD88xTRIF KO MEF | This paper | Primary murine embryonic fibroblasts. See Materials and methods, Section 2 | |
| Other | TBK1 KO MEF | This paper | Primary murine embryonic fibroblasts. See Materials and methods, Section 2 | |
| Other | TBK1 HET MEF | This paper | Primary murine embryonic fibroblasts. See Materials and methods, Section 2 | |
| Other | STING GT MEF | This paper | Primary murine embryonic fibroblasts. See Materials and methods, Section 2 | |
| Other | MAVS (IPS1) KO MEF | This paper | Primary murine embryonic fibroblasts. See Materials and methods, Section 2 | |
| Other | MAVS (IPS1) KO MEF | This paper | Primary murine embryonic fibroblasts. See Materials and methods, Section 2 | |
| Other | MAVS (IPS1) KO MEF | This paper | Primary murine embryonic fibroblasts. See Materials and methods, Section 2 | |
| Other | ZBP1 KO MEF | This paper | Primary murine embryonic fibroblasts. See Materials and methods, Section 2 | |
| Other | DNA-PKSCID MEF | This paper | Primary murine embryonic fibroblasts. See Materials and methods, Section 2 | |
| Other | cGAS KO MEF | This paper | Primary murine embryonic fibroblasts. See Materials and methods, Section 2 | |
| Virus (murine cytomegalovirus) | MCMV WT1 | Cheng et al., 2010 | ||
| Virus (murine cytomegalovirus) | MCMV GFP | Henry et al., 2000 | ||
| Sequence-based reagent | MCMV IE1 | IDT DNA | TAQman assay | Forward: 5’-CCCTCTCCTAACTCTCCCTTT-3’; Reverse: 5’-TGGTGCTCTTTTCCCGTG −3’; Probe: 5’- TCTCTTGCCCCGTCCTGAAAACC-3’ |
| Sequence-based reagent | ACTB | IDT DNA | TAQman assay | Forward: 5’-AGCTCATTGTAGAAGGTGTGG-3’; Reverse: 5’- GGTGGGAATGGGTCAGAAG-3’; Probe: 5’-TTCAGGGTCAGGATACCTCTCTTGCT-3’ |
| Sequence-based reagent | IFNB1 | Thermo Fisher Scientific | TAQman assay | Mm00439546_s1 |
| Sequence-based reagent | (pan)Ifna | IDT DNA | TAQman assay | Forward: 5’-CTTCCACAGGATCACTGTGTACCT-3’; Reverse: 5’-TTCTGCTC TGACCACCTCCC-3’; Probe: 5’-AGAGAGAAGAAACACAGCCC CTGTGCC-3’ |
| Sequence-based reagent | GAPDH | Thermo Fisher Scientific | TAQman assay | Mm99999915_g1 |
| Antibody | Anti-mouse NK1.1 PE-Cy7 (Mouse monoclonal) | Thermo Fisher Scientific | Cat#: 25-5941-82; RRID:AB_469665 | FACS (1:100) |
| Antibody | Anti-mouse NKp46 PerCP-eFluor710 (Rat monoclonal) | Thermo Fisher Scientific | Cat#: 46-3351-82; RRID:AB_1834441 | FACS (1:100) |
| Antibody | Anti-mouse CD3 APC-eFluor780 (Armenian hamster monoclonal) | Thermo Fisher Scientific | cat# 47-0031-82, RRID:AB_11149861 | FACS (1:100) |
| Antibody | Anti-mouse CD19 APC-eFluor780 (Rat monoclonal) | Thermo Fisher Scientific | Cat# 47-0193-82, RRID:AB_10853189 | FACS (1:100) |
| Antibody | Ly49H FITC (Mouse monoclonal) | Made in-house | FACS (1:200) | |
| Antibody | Anti-mouse CD31 PE (Rat monoclonal) | Thermo Fisher Scientific | Cat# 12-0311-83, RRID:AB_465633 | FACS (1:100) |
| Antibody | Anti-mouse PDCA1 PE (Mouse monoclonal) | Thermo Fisher Scientific | Cat# 12-3171-81, RRID:AB_763427 | FACS (1:50) |
| Antibody | Anti-mouse gp38 PE-Cy7 (Syrian hamster monoclonal) | Thermo Fisher Scientific | Cat# 25-5381-82, RRID:AB_2573460) | FACS (1:100) |
| Antibody | Anti-mouse CD45 APC (Rat monoclonal) | Thermo Fisher Scientific | Cat# 17-0451-83, RRID:AB_469393) | FACS (1:50) |
| Antibody | Anti-mouse CD11c APC-eFluor780 (Armenian hamster monoclonal) | Thermo Fisher Scientific | Cat# 47-0114-82, RRID:AB_1548652) | FACS (1:50) |
| Antibody | Anti-mouse Ly49H APC (Mouse monoclonal) | Thermo Fisher Scientific | Cat# 17-5886-82, RRID:AB_10598809 | FACS (1:100) |
| Antibody | Anti-mouse Perforin PE (Rat monoclonal) | Thermo Fisher Scientific | Cat# 12-9392-82, RRID:AB_466243 | FACS (1:50) |
| Antibody | Anti-mouse Granzyme B APC (Mouse monoclonal) | Thermo Fisher Scientific | Cat# MHGB05, RRID:AB_10373420 | FACS (1:100) |
| Antibody | Anti-mouse IFNg eFluor450 (Rat monoclonal) | Thermo Fisher Scientific | Cat# 48-7311-82, RRID:AB_1834366 | FACS (1:100) |
| Commercial assay or kit | Mouse IFNB ELISA | Biolegend | 439407 | |
| Commercial assay or kit | Cytofix/Cytoperm kit | BD Biosciences | 554714 | |
| Software, algorithm | Prism | Graphpad | RRID: SCR_002798 | |
| Software, algorithm | Flowjo | Treestar Inc | RRID:SCR_008520 | |
| Other | Viability stain eFluor 506 | Thermofisher Scientific | 65-0866-14 | FACS (1:1000) |





