Genetic variant in 3’ untranslated region of the mouse pycard gene regulates inflammasome activity
Figures
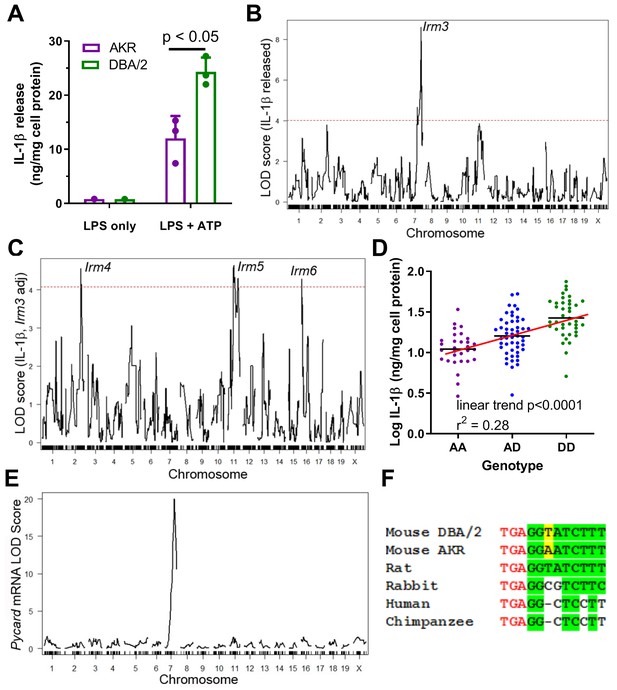
Strain effect on IL-1β release associated with Pycard gene.
(A) IL-1β released into conditioned media (normalized to cell protein) from AKR (magenta) or DBA/2 (green) BMDM after treatment with LPS (4 hr, single well) only or LPS (4 hr)+ATP (30 min, biological triplicates), p<0.05 where indicated by two-tailed t-test, mean and SD shown. Representative of several independent experiments. (B) QTL mapping for IL-1β release from F4 BMDMs, showing the prominent Irm3 QTL peak on chromosome 7. (C) QTL mapping for IL-1β release from F4 BMDMs after adjusting for the effect of the Irm3 QTL, showing Irm4-6 QTLs on chromosomes 2, 11, and 16, respectively (dashed red line in (B and C) represents the genome-wide significance threshold). (D) Log10 IL-1β release from F4 BMDMs by genotype at the strongest associated SNP at the Irm3 peak (AA, AKR homozygotes; AD heterozygotes; DD, DBA/2 homozygotes; p<0.0001 by ANOVA linear trend posttest). (E) eQTL mapping for Pycard mRNA levels, showing the cis-eQTL peak mapping to chromosome seven where the Pycard gene resides. (F) Pycard gene sequence conservation at the stop codon (red text) and the immediate 3’UTR (conserved residues highlighted in green and the AKR-DBA/2 SNP in yellow). Source data in file Source data 1.
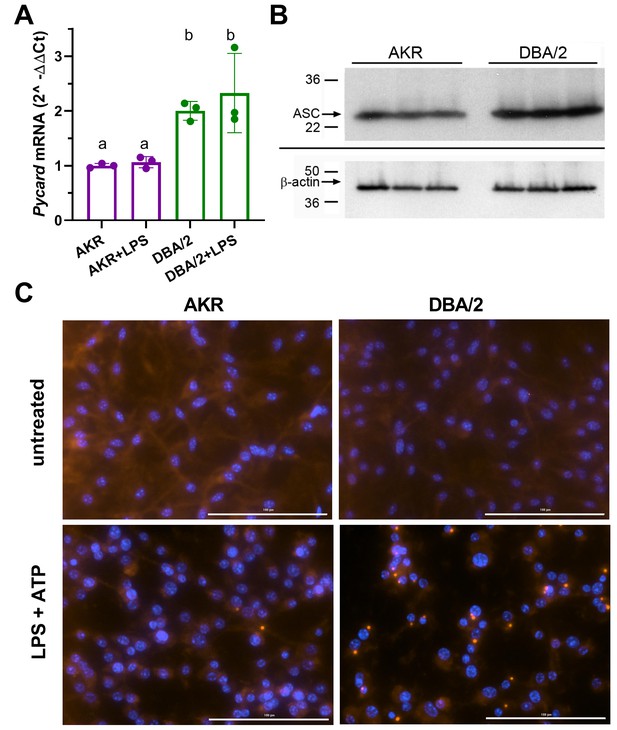
Strain effects on Pycard/ACS expression and inflammasome speck formation.
(A) Relative Pycard mRNA levels in AKR (magenta) and DBA/2 (green) BMDM, showing no induction by LPS (different letters above columns show p<0.05 by ANOVA Tukey posttest, mean and SD shown). Median of technical triplicates of biological triplicates plotted, representative of two independent experiments. (B) Western blot for ASC (top) and β-actin (bottom) in biological triplicate lysates from AKR and DBA/2 BMDM. Densitometric analysis revealed a 50% increase in the ASC/β-actin ratio (p<0.01 by two-tailed t-test). (C) Immunofluorescent staining for ACS specks (red) showing assembled inflammasomes and nuclei (blue) in AKR and DBA/2 BMDM with or without inflammasome priming and activation by LPS (4 hr)+ATP (30 min) treatment. For the fields shown in the lower panels, primed and activated AKR BMDM yielded 2 specks among 98 nuclei, while DBA/2 BMDM yielded 23 specks among 58 nuclei. Source data in file Source data 1. Unedited western blots in (B) unedited western blot source data.docx.
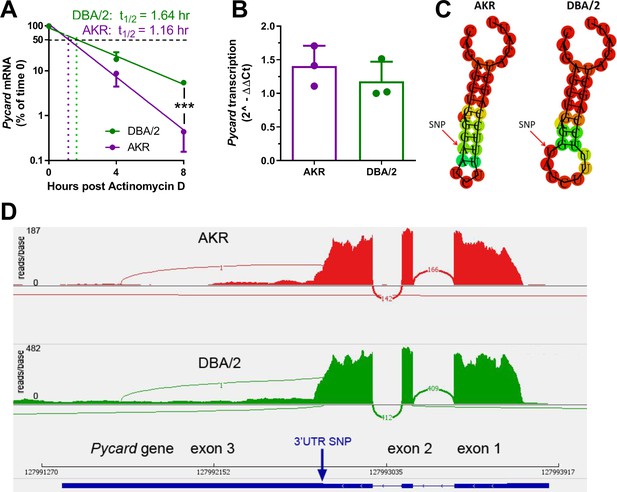
Strain effect on Pycard mRNA turnover and structure.
(A) Semi-log plot of Pycard mRNA turnover after Actinomycin D treatment of AKR (magenta) and DBA/2 (green) BMDM (***, p<0.001 by two-tailed t-test). Each point is the mean ± SD of biological triplicates using the mean of technical triplicates. (B) Relative level of Pycard mRNA run-on transcription in AKR (magenta) and DBA/2 (green) BMDM (not significant by two-tailed t-test). Biological triplicates, mean and SD shown. (C) Predicted structure of AKR and DBA/2 Pycard mRNA segments near the 3’UTR SNP. (D) Sashimi plot of exon junctional reads and read depth histogram (IGV browser view) of RNAseq from AKR and DBA/2 BMDM, with the Pycard gene exon-intron structure below (gene on lower strand, 5’ to 3’ from right to left). Source data in file Source data 1.
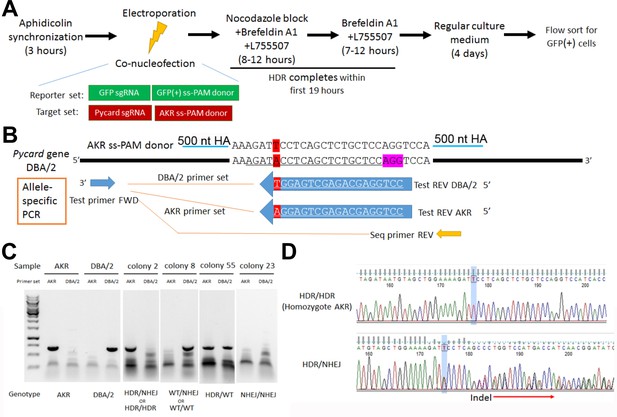
Pycard gene editing of ES cells to convert the DBA/2 allele to the AKR allele.
(A) Strategy used to decrease NHEJ by use of HDR reporter and small molecules to modulate cell cycle and inhibit NHEJ. (B) Sequence of the AKR allele ss donor (3’UTR SNP highlighted in red) with 500 nt homology arm (HA), which will change the SNP from DBA/2 to AKR and eliminate Cas9 re-cutting, since the SNP is within the sgRNA sequence (underlined in the DBA/2 gene sequence). The sequence of the AKR and DBA/2 allele-specific PCR reverse primers are also shown with the SNP at the 3’ end, along with the positions of the common forward primer and the reverse PCR primer used for sequencing the edited clonally derived genomic DNA. (C) Example of allele-specific PCR using AKR and DBA/2 genomic controls, and DNA from expanded colonies after gene editing. Genotypes cannot all be distinguished, as one of both alleles may be edited by NHEJ precluding DNA amplification. (D) Sanger sequencing of DNA after gene editing showing a homozygous HDR conversion to the AKR allele (top) and a compound heterozygous with editing to one AKR allele and one indel allele due to NHEJ (bottom).
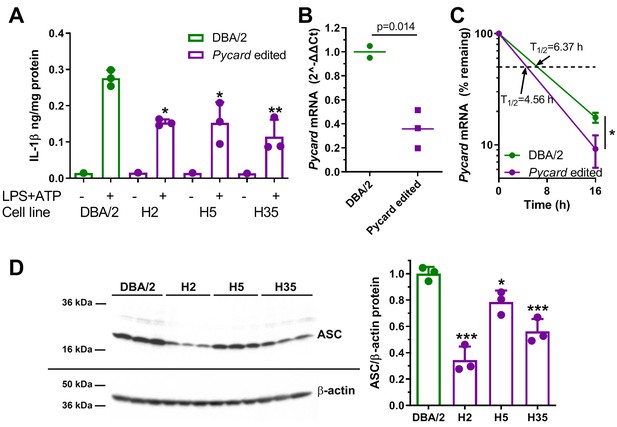
Gene editing alters IL-1β release and Pycard expression in ESDM.
(A) IL-1β release from ESDM derived from DBA/2 ES (green) and three independent homozygous Pycard edited ES lines (magenta), in the absence or presence of inflammasome priming and activation with LPS +ATP (each point is a biological replicate; *, p<0.05; **, p<0.01 vs DBA/2 derived ESDM in the presence of LPS +ATP by ANOVA with Dunnett's multiple comparisons test, mean and SD shown). (B) Pycard mRNA levels in two DBA/2 ESDM differentiations and three independent homozygous Pycard edited ESDM (p=0.014 by two-tailed t-test). Each point is a biological replicate of qPCR technical triplicates, mean shown. (C) Semi-log plot of Pycard mRNA turnover after Actinomycin D treatment of ESDM derived from two differentiations of DBA/2 ES and three independent homozygous Pycard edited ES lines (magenta), (*, p<0.05 by two-tailed t-test). Each point is the average of biological triplicates of qPCR technical triplicates, mean and SD shown. (D) Left side, western blot for ASC (top) and β-actin (bottom) from ESDM lysates derived from DBA/2 ES and three independent homozygous Pycard edited ES lines. Right side, densitometry of western blot showing ASC levels are lower in all three Pycard edited cell lines (*, p<0.05; ***, p<0.001 vs. DBA/2 derived ESDM by ANOVA with Dunnett's multiple comparisons test, mean and SD shown).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Pycard | Ensemble | ENSG00000103490 | |
| Strain, strain background (Mus musculus) | AKR/J | JAX | 648 | |
| Strain, strain background (Mus musculus) | DBA/2J | JAX | 671 | |
| Cell line (Mus musculus) | DBA/2J mouse ES cell line AC173/GrsrJ | JAX | 000671C02 | |
| Cell line (Mus musculus) | (Puromycin-resistant MEF feeder cells) | Cell Biolabs | CBA-312 | |
| Cell line (Mus musculus) | Neomycin-resistant MEF feeder cells (Cell Biolabs, CBA-311) | Cell Biolabs | CBA-311 | |
| Transfected construct (S. pyogenes) | Cas9 expression plasmid pSpCas9(BB)−2A-Puro | Addgene | PX459 | |
| Transfected construct (Aequorea Victoria) | MSCV-miRE-shRNA IFT88-PGK-neo-IRES-GFP plasmid, | Addgene | 73576 | |
| Antibody | Mouse monoclonal anti Cas9 (Diagenode, C15200203) | Diagenode | C15200203 | IHC (1:1000) |
| Antibody | Rabbit monoclonal anti-mouse AS | Cell Signalling | 67824 | WB(1:500) |
| Antibody | Rabbit polyclonal anti-ASC N-terminus | Santa Cruz Biotech | Sc-22514-R | IF (10 μg/ml) |
| Antibody | Goat polyclonal alexa Flour 568 anti-rabbit IgG | ThermoFisher | A-11011 | IF (2 μg/ml) |
| Antibody | Mouse monoclonalHRP-conjugated anti β-actin | Santa Cruz Biotech | Sc-47778-HRP | (WB (1:20,000)) |
| Sequence-based reagent | qPCR for mouse Pycard | ThermoFisher | 4331182, Assay ID: Mm00445747_g1 | |
| Sequence-based reagent | qPCR for mouse Actb | ThermoFisher | 4448484, Assay ID: Mm02619580_g1 | |
| Sequence-based reagent | Nuclear run-on qPCR for mouse Pycard | ThermoFisher | 4441114, Assay ID: AJMSHN7 | |
| Sequence-based reagent | sgRNA for GFP stop codon | Synthego | Custom | GGGCGAGGGCGAUGCCACCU |
| Sequence-based reagent | sgRNA for Pycard 3’ UTR | Synthego | Custom | AGAUACCUCAGCUCUGCUCC |
| Peptide, recombinant protein | Leukaemia inhibitory factor | Millipore-Sigma | ESG1107 | |
| Peptide, recombinant protein | Mouse IL-3 | R and D Systems | 403 ML | |
| Commercial assay or kit | Mouse Il-1β ELISA | R and D Systems | MLB00C | |
| Commercial assay or kit | SuperScript VILO cDNA synthesis kit | ThermoFisher | 1175505 | |
| Commercial assay or kit | iScript cDNA synthesis kit | BioRad | 1708891 | |
| Chemical compound, drug | LPS from E. coli O55:B5 | Sigma | L6529 | |
| Chemical compound, drug | ATP | Sigma | A2383 | |
| Software, algorithm | Custom special R functions | This paper | https://github.com/BrianRitchey/qtl | See Methods section |
| Software, algorithm | r/QTL | Reference 30 in this paper | https://rqtl.org/download/ | |
| Other | 30 µm sterile filters | Sysmex | 04-004-2326 | To filter out embryonic bodies during ESDM differentiation |
Additional files
-
Source data 1
Source data in file Figure 5.
Unedited western blots in Figure 5D unedited western blot source data.docx.
- https://cdn.elifesciences.org/articles/68203/elife-68203-data1-v1.zip
-
Supplementary file 1
Supplementary file.
(a) Genes within the Irm3 QTL interval. All genes in the interval are shown along with the Mb position on chromosome 7. Other columns show distance from the LOD peak in Mb, whether a cis-eQTL was found and if so the LOD score for the eQTL, PubMed hits for the gene and the terns ‘inflammasome’ and ‘IL-1b’, the number of non-synonymous SNPs between the AKR and DBA/2 mouse strains, and the PROVEAN analysis to determine the number of non-synonymous SNPs likely to be deleterious to protein function. a, LOD score for cis-eQTL based on our prior BMDM strain intercross (Reference: J Hsu and JD Smith, PMID: 23525445 DOI: 10.1161/JAHA.112.005421); b, total number of PubMed hits for Boolean queries of the respective gene name and terms of interest.; c, non-synonymous SNPs between AKR and DBA/2 mice; d, PROVEAN, number of SNPs predicted to be deleterious by PROVEAN software. Yellow highlighting, top candidate gene. (b) Pycard gene sequencing PCR primer pairs. The sequence of the 6 PCR primer pairs used for Sanger sequencing of the Pycard gene in AKR and DBA/2 genomic DNA, along with the position of the primers relative to the transcription start site (TSS).
- https://cdn.elifesciences.org/articles/68203/elife-68203-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68203/elife-68203-transrepform-v1.docx





