circFL-seq reveals full-length circular RNAs with rolling circular reverse transcription and nanopore sequencing
Figures
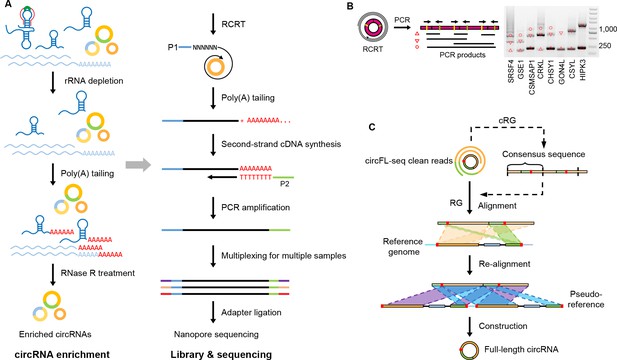
Diagram of circFL-seq workflow.
(A) Experimental operation of circFL-seq consisted of circRNA enrichment, library construction, and nanopore sequencing. (B) PCR validation of rolling circle products from the circFL-seq cDNA library. The yellow and green lines indicate the positions of the PCR primers. The upward triangle, downward triangle, and circle symbols denote the 0-circle, 1-circle, and 2-circle cDNA products. (C) Computational pipeline of circFL-seq. circFL-seq clean reads were directly used in RG mode or were self-corrected for consensus sequences in cRG mode to reconstruct full-length circRNAs. circRNA, circular RNA.
-
Figure 1—source data 1
Original figures of gels.
This file includes figures with uncropped gels.
- https://cdn.elifesciences.org/articles/69457/elife-69457-fig1-data1-v2.zip
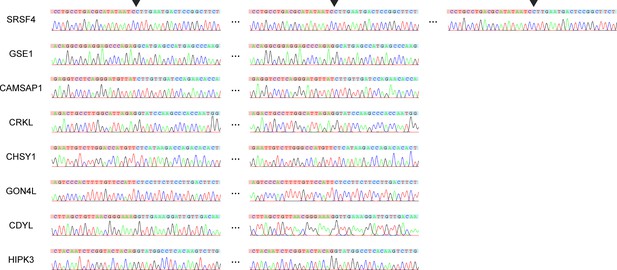
Sanger sequencing of rolling circular bands.
PCR was performed with the circFL-seq library of HEK293T cells as a template.

Analysis of full-length circRNA in eight samples.
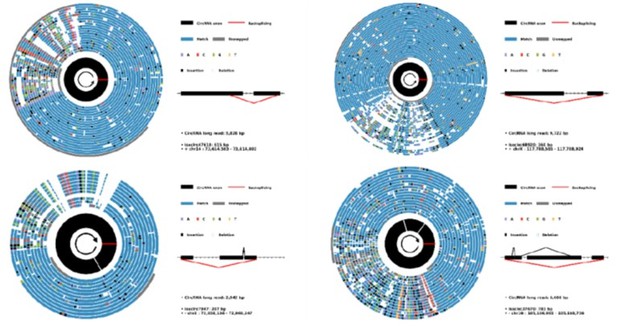
(A) Stacked bar plot represents the number of full-length circRNA isoforms detected by RG and cRG for six cell lines. (B) Expression correlation matrix for circRNA BSJs and isoforms of all samples. The color scale corresponds to Pearson’s correlation coefficient. (C) Stacked bar plot represents the number of circRNA isoforms with read counts≥5 from known or novel BSJs based on the circRNA database. (D) Boxplot showing the length distribution per isoform for circRNA isoforms with read counts≥5 in all samples. Box lefts or rights are lower or upper quartiles, the bar is the median, and whiskers are the median±1.5×interquartile range. (E) Stacked bar plot showing the fraction of exon numbers per isoform for circRNA isoforms with read counts≥5 in all samples. (F) Boxplot showing the length distribution per exon for circRNA isoforms with read counts≥5 in all samples. Box bottoms or tops are lower or upper quartiles, the bar is the median, and the whiskers are the median±1.5×interquartile range. (G) Diagram of four types of alternative splicing (AS) events in circRNA: exon skipping (ES), alternative 3′ splice site (A3SS), alternative 5′ splice site (A5SS), and intron retention (IR). (H) Plot showing the coverage of full-length circRNA reads in the position of CDR1as for circFL-seq data of six cell lines (replicate data were merged). Structures of the two isoforms of CDR1as are shown at the bottom. (I–K) AS events (one ES and one IR) of circ-TMEM138 detected by circFL-seq (I), agarose gel electrophoresis (J), and Sanger sequencing (K). Red/blue arcs are forward/reverse primers for validation of back-splicing junctions (BSJs) and forward splicing junctions (FSJs). Asterisks denote FSJs. Downward triangles denote BSJs. BSJ, back-splicing junction; circRNA, circular RNA; RG, reference guide.

Clean read distribution of circFL-seq data of six cell lines.
(A) Histograms showing the distribution of clean reads (blue) and full-length circRNA reads (red) for each sample. The percentages represent the circRNA read proportion in clean reads. (B) Histograms showing the distribution of full-length circRNA read amounts. circRNA, circular RNA.
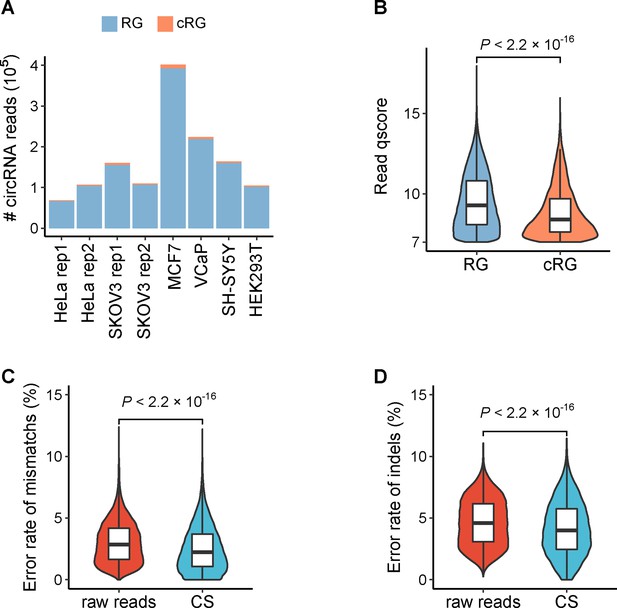
CircRNA reads identified from circFL-seq data of six cell lines.
(A) Stacked bar plot representing the number of full-length circRNA isoforms detected by RG and cRG for eight samples. (B) Boxplot showing the read qscore distribution of circRNA isoforms of all samples. The qscore representing the read quality was extracted from the sequencing summary file. Boxplot showing the error rate of mismatches (C) and indels (D) for raw reads and consensus sequence (CS) of cRG-identified circRNA isoforms. circRNA, circular RNA; RG, reference guide.

Scatter plot showing the correlation of circRNA at the BSJ level (A, B) and isoform level (C, D) between circFL-seq replicates.
CircRNA BSJs/isoforms with read counts>0 in at least one replicate were included. BSJ, back-splicing junction; circRNA, circular RNA.
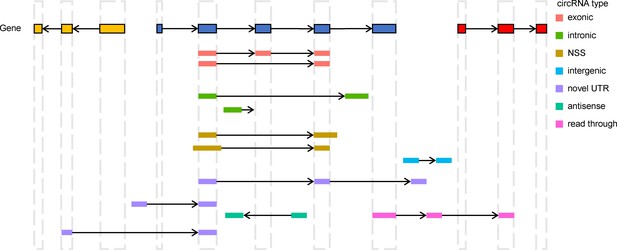
Diagram of circRNA types.
The classification is based on the positions of BSJs and boundary exons following the principles below. Exonic: the circRNA body is totally located inside of one gene from the same strand. Both of the BSJs are identical to annotated junctions. Intronic: the circRNA body is also totally located inside one gene from the same strand. However, at least one of the boundary exons does not overlap with any annotated exon. Novel splicing site (NSS): the circRNA body is also totally located inside one gene from the same strand. However, both boundary exons overlap with annotated exons, with at least one BSJ different from the annotated linear junction. Intergenic: the whole body of circRNA is located in an intergenic region. Novel UTR: the circRNA body partially overlaps with only one gene from the same strand, and at least one BSJ is located in an intergenic region. Antisense: there is no overlap between circRNA and any gene from the same strand. However, the circRNA overlaps gene(s) from the antisense strand. Read-through: BSJs are located in different genes with the same strand. BSJ, back-splicing junction; circRNA, circular RNA.
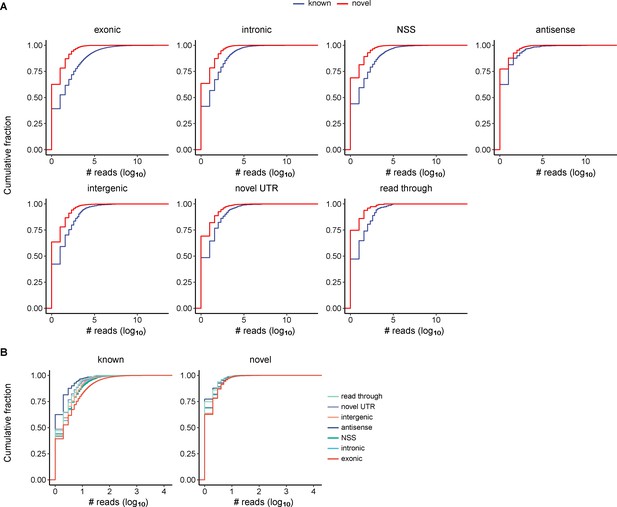
Cumulative distribution of read counts for circRNA isoforms identified by circFL-seq from six cell lines.
CircRNA isoforms were classified based on database status and annotation types. (A) For each annotation type (exonic, intronic, NSS, intergenic, novel UTR, antisense, and read-through), the cumulative distribution of read counts is classified to known and novel status based on whether their BSJs are annotated in the circRNA database. (B) For known and novel status, the cumulative distribution of read counts is classified into seven annotation types. BSJ, back-splicing junction; circRNA, circular RNA.
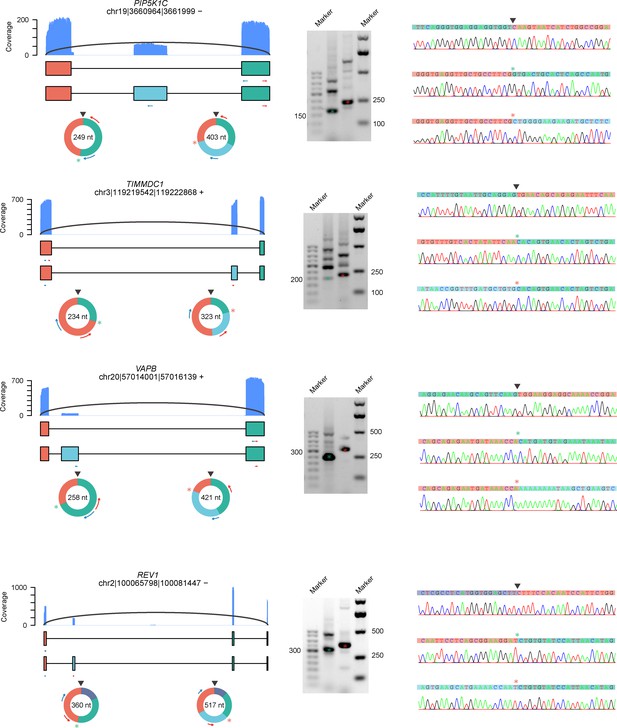
CircRNAs with exon skipping validated by RT-PCR and Sanger sequencing in HeLa cells.
RT-PCR was performed with RNase R-treated RNA. The coverage of full-length circRNA reads mapped to the reference genome is shown. circRNA, circular RNA.
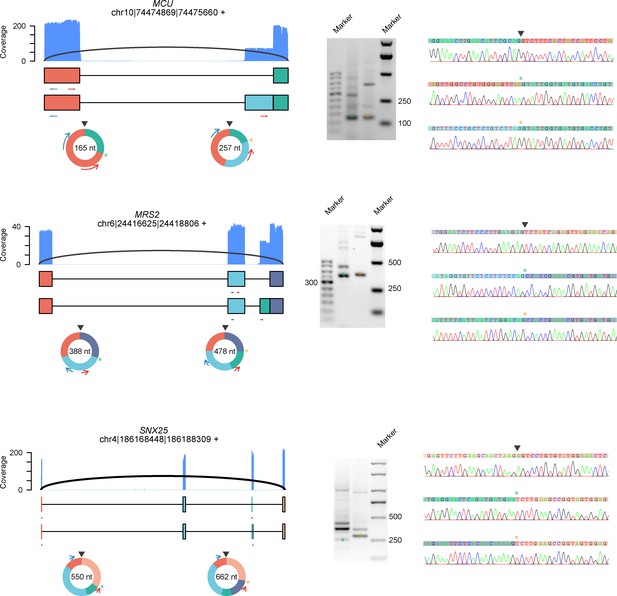
CircRNAs with alternative 3′/5′ splicing sites (A3SS for circRNAs from MCU and MRS2, A5SS for circRNA from SNX25) were validated by RT-PCR and Sanger sequencing in HeLa cells.
RT-PCR was performed with RNase R-treated RNA. The coverage of full-length circRNA reads mapped to the reference genome is shown. A3SS, alternative 3′ splice site; alternative 5′ splice site; circRNA, circular RNA.

CircRNAs with intron retention validated by RT-PCR and Sanger sequencing in HeLa cells.
RT-PCR was performed with RNase R-treated RNA. The coverage of full-length circRNA reads mapped to the reference genome is shown. circRNA, circular RNA.
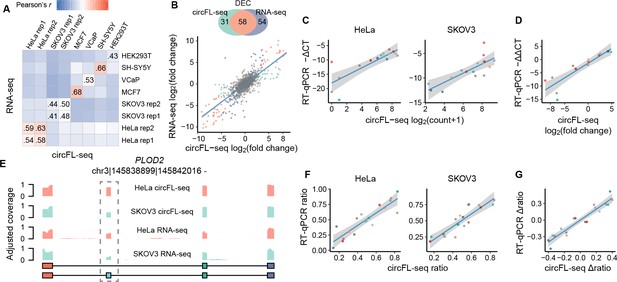
Quantification of circRNA at the BSJ and isoform levels.
(A) Expression correlation matrix of circRNA BSJ quantified by circFL-seq and RNA-seq for six cell lines. The numbers in the matrix represent Pearson’s correlation coefficients. (B) Comparison of differentially expressed circRNA (DEC) detection between circFL-seq and RNA-seq. Top panel: Venn diagram showing the number of DECs detected by circFL-seq (green), RNA-seq (purple), and both methods (orange). Bottom panel: scatter plot showing the correlation of fold change (log base 2) for HeLa and SKOV3 cells between circFL-seq and RNA-seq. (C) Scatter plot showing the correlation of the expression levels of 16 circRNA BSJs for HeLa (left) and SKOV3 (right) cells between circFL-seq and RT-qPCR. (D) Scatter plot showing the correlation of fold changes (log base 2) of the 16 BSJs for HeLa and SKOV3 cells between circFL-seq and RT-qPCR. (E) Plot showing the adjusted coverage of full-length circRNA reads and RNA-seq reads in the position of circRNA from PLOD2. The circular structures of the two circRNA isoforms are shown in the lower panel. (F) Scatter plot showing the correlation of the transcript ratio of 18 circRNA isoforms from nine circRNA BSJs (each BSJ has two isoforms) for HeLa (left) and SKOV3 (right) cells between circFL-seq and RT-qPCR. The relative expression of target BSJs/isoforms quantified by RT-qPCR was determined with RNase R-treated samples and GAPDH from total RNA without RNase R treatment as a reference. (G) Scatter plot showing the correlation of the differential ratio (∆ratio) of the 18 isoforms for HeLa and SKOV3 cells between circFL-seq and RT-qPCR. The shaded areas denote 95 % confidence intervals. BSJ, back-splicing junction; circRNA, circular RNA.
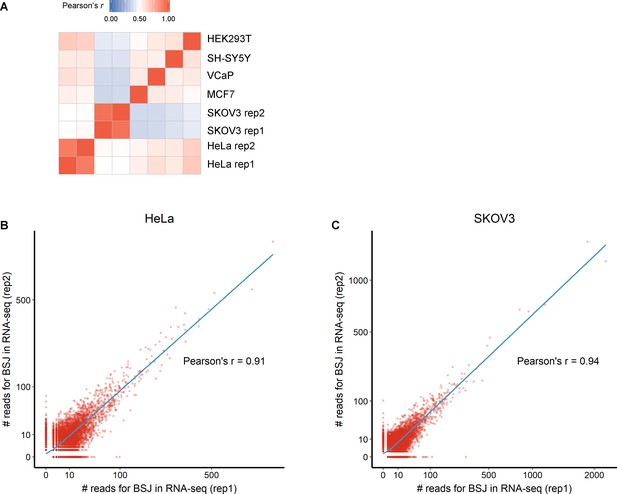
Correlations of circRNA BSJs among RNA-seq samples from six cell lines.
(A) Expression correlation matrix for circRNA BSJs among six cell lines. The color scale corresponds to Pearson’s correlation coefficients. Scatter plot showing the correlations of BSJs between HeLa (B) or SKOV3 (C) replicates. CircRNA BSJs with read counts > 0 in at least one replicate were included.
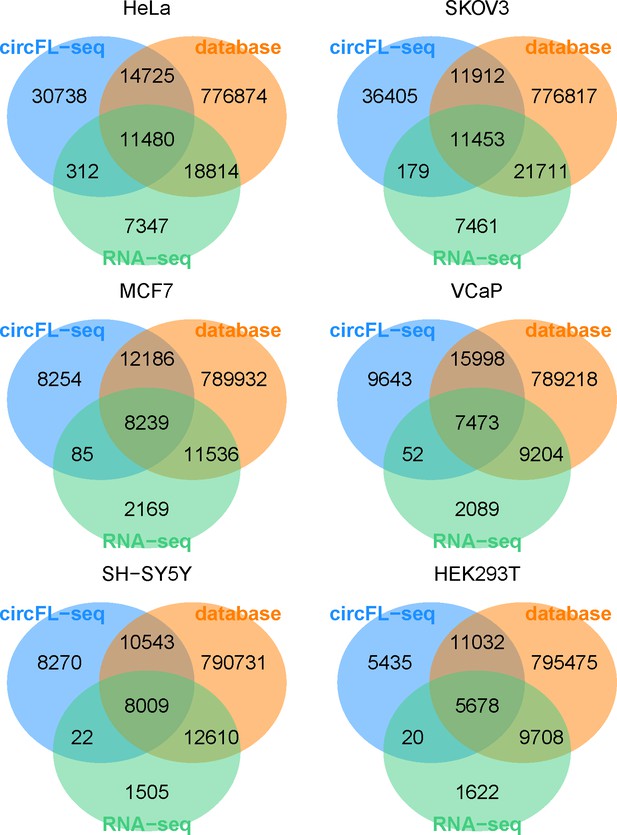
Venn diagram of BSJs detected by circFL-seq, RNA-seq, and database.
BSJ, back-splicing junction.

CircRNA read distribution of eight samples of six cell lines.
Bar plot showing the distribution of known or novel circRNA BSJs with different read counts as the threshold for circFL-seq (A) and RNA-seq (B) data. BSJ, back-splicing junction; circRNA, circular RNA.

Comparison of circFL-seq and RNA-seq for length of full-length circRNA of six cell lines.
For RNA-seq, full-length circRNAs were reconstructed by CIRI-full. circRNA, circular RNA.
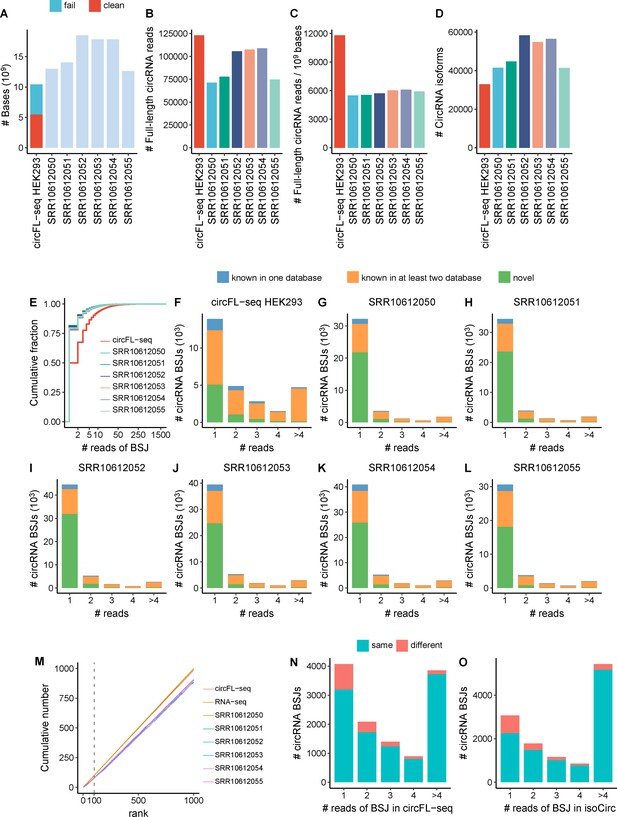
Comparison of circFL-seq and isoCirc for full-length circRNA detection in the HEK293 cell line.
(A) Stacked bar plot showing the number of sequenced bases. circFL-seq includes fail and clean bases. Fail bases are from low-quality reads with qscore<7 and trimmed adapters. (B) Bar plot showing the number of full-length circRNA reads. (C) Bar plot showing full-length circRNA reads per 109 raw sequenced bases. (D) Bar plot showing the number of full-length circRNA isoforms. (E) Cumulative distribution of read counts of BSJs. (F–L) Stacked bar plot showing the distribution of known or novel circRNA BSJs for different read counts. (M) Plot showing the cumulative number of top expressed circRNAs of circFL-seq, isoCirc, and RNA-seq detected in the database. Stacked bar plot showing the distribution of read counts of common circRNA BSJs detected in circFL-seq (N) and isoCirc (O). ‘Same’ and ‘different’ represent BSJs w/wo the same isoforms between circFL-seq and isoCirc. The isoCirc analysis in (N, O) combines circRNA results from all six HEK293 isoCirc libraries. BSJ, back-splicing junction; circRNA, circular RNA.

Scatter plot showing the correlation of circRNA BSJs between circFL-seq and RNA-seq samples of six cell lines.
BSJ, back-splicing junction; circRNA, circular RNA.
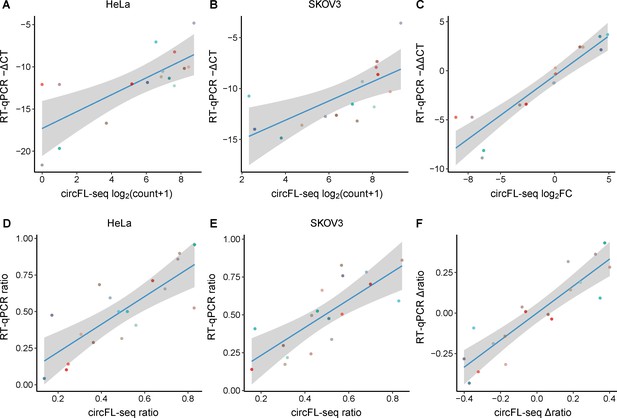
Evaluation of circRNA quantification between circFL-seq and RT-qPCR.
The relative expression of target BSJs and isoforms quantified by RT-qPCR was performed with samples without RNase R treatment.GAPDH from total RNA without RNase R treatment was used as a reference. (A, B) Scatter plot showing the correlation of expression levels of 16 circRNA BSJs for HeLa (A) and SKOV3 (B) cells between circFL-seq and RT-qPCR. (C). Scatter plot showing the correlation of fold change (log base 2) of the 16 BSJs for HeLa and SKOV3 cells between circFL-seq and RT-qPCR. (D, E) Scatter plot showing the correlation of the transcript ratio of 18 circRNA isoforms from 9 circRNA BSJs (each BSJ has two isoforms) for HeLa (D) and SKOV3 (E) cells between circFL-seq and RT-qPCR. (F) Scatter plot showing the correlation of the differential ratio (∆ratio) of the 18 isoforms for HeLa and SKOV3 cells between circFL-seq and RT-qPCR. BSJ, back-splicing junction; circRNA, circular RNA.
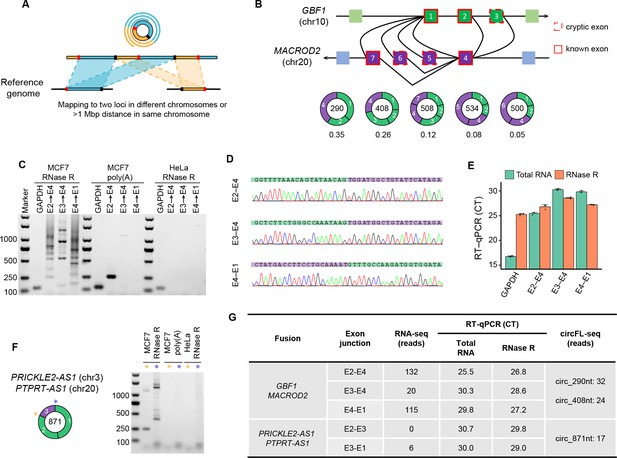
Detection and validation of fusion circRNA (f-circRNA) in the MCF7 cell line.
(A) Diagram of identification of f-circRNA with circFL-seq data. (B) Diagram of five high-quality f-circRNA isoforms (read counts≥5) fused by GBF1 and MACROD2. The transcript ratio represents the fractions of the isoforms. (C–E) Validation of f-circRNA junctions from GBF1/MACROD2 by agarose gel electrophoresis (C), Sanger sequencing (D), and RT-qPCR (E). (C) Agarose gel electrophoresis showing the RT-PCR products of f-circRNA junctions with RNase R-treated MCF7 and HeLa RNA and poly(A) selected MCF7 RNA as a template. (F) Agarose gel electrophoresis showing the RT-PCR products of f-circRNA junctions from PRICKLE2-AS1/PTPRT-AS1. (G) Information on five f-circRNA junctions detected by circFL-seq, RNA-seq, and RT-qPCR.

Sanger validation of sequences of f-circRNA from GBF1/MACROD2.
The forward and reverse primers are highlighted. f-circRNA, fusion circRNA.

Sanger validation of the sequence of f-circRNA from PRICKLE2-AS1/PTPRT-AS1.
The forward and reverse primers are highlighted. f-circRNA, fusion circRNA.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HeLa | Jiadong Wang Laboratory | RRID:CVCL_0030 | |
| Cell line (H. sapiens) | SKOV3 | Jiadong Wang Laboratory | RRID:CVCL_0532 | |
| Cell line (H. sapiens) | MCF7 | Jiadong Wang Laboratory | RRID:CVCL_0031 | |
| Cell line (H. sapiens) | HEK293T | Jiadong Wang Laboratory | RRID:CVCL_0063 | |
| Cell line (H. sapiens) | SH-SY5Y | Jian Chen Laboratory | RRID:CVCL_0019 | |
| Cell line (H. sapiens) | VCaP | iCell Bioscience | RRID:CVCL_2235 | |
| Cell line (H. sapiens) | HEK293 | iCell Bioscience | RRID:CVCL_0045 | |
| Commercial assay or kit | Total RNA of human brain | Clontech | Cat. #: 636530 | |
| Commercial assay or kit | Total RNA of human testis | Clontech | Cat. #: 636533 |
Summary of confounding factors among three methods.
| details | circFL-seq | CIRI-long | isoCirc |
|---|---|---|---|
| input of total RNA (μg) | 2 | 1 | 20 |
| species | Human | mouse | human |
| samples | 7 cell linesbrain and testis | brain | 1 cell line12 tissues |
| platform | ONT PromethION, MinION | ONT MinION | ONT MinION |
| libraries per sample | one/two | multiple | multiple |
| libraries per Flow Cell (sequencing depth) | one/multiple | multiple | one |
Comparison of isoCirc and circFL-seq for BSJ detection in HEK293 cell line.
| total circRNA BSJs | ||||||
|---|---|---|---|---|---|---|
| # read counts for BSJ | 1 | 2 | 3 | 4 | >4 | all |
| isoCirc HEK293 SRR10612050 | 32,204 | 3,572 | 1,237 | 620 | 1,777 | 39,410 |
| isoCirc HEK293 SRR10612051 | 34,493 | 3,916 | 1,361 | 687 | 1,915 | 42,372 |
| isoCirc HEK293 SRR10612052 | 44,586 | 5,270 | 1,707 | 860 | 2,635 | 55,058 |
| isoCirc HEK293 SRR10612053 | 39,484 | 5,274 | 1,871 | 1,022 | 2,970 | 50,621 |
| isoCirc HEK293 SRR10612054 | 40,928 | 5,259 | 1,897 | 1,071 | 3,009 | 52,164 |
| isoCirc HEK293 SRR10612055 | 30,647 | 3,779 | 1,387 | 710 | 2,024 | 38,547 |
| isoCirc HEK293 all | 158,875 | 23,302 | 8,821 | 5,133 | 26,782 | 222,913 |
| circFL-seq HEK293 | 13,906 | 4,889 | 2,830 | 1,525 | 4,719 | 27,869 |
| known circRNA BSJs annotated in database | ||||||
| # read counts for BSJ | 1 | 2 | 3 | 4 | >4 | all |
| isoCirc HEK293 SRR10612050 | 10,458 | 2,528 | 1,080 | 571 | 1,620 | 16,257 |
| isoCirc HEK293 SRR10612051 | 10,889 | 2,663 | 1,194 | 615 | 1,751 | 17,112 |
| isoCirc HEK293 SRR10612052 | 12,727 | 3,447 | 1,451 | 773 | 2,396 | 20,794 |
| isoCirc HEK293 SRR10612053 | 14,828 | 3,893 | 1,665 | 944 | 2,711 | 24,041 |
| isoCirc HEK293 SRR10612054 | 15,078 | 3,834 | 1,678 | 971 | 2,750 | 24,311 |
| isoCirc HEK293 SRR10612055 | 12,534 | 2,969 | 1,264 | 660 | 1,860 | 19,287 |
| isoCirc HEK293 all | 28,917 | 12,088 | 6,821 | 4,411 | 25,301 | 77,538 |
| circFL-seq HEK293 | 8,836 | 3,821 | 2,377 | 1,365 | 4,589 | 20,988 |
| % known circRNA BSJs | ||||||
| # read counts for BSJ | 1 | 2 | 3 | 4 | >4 | all |
| isoCirc HEK293 SRR10612050 | 32.5 | 70.8 | 87.3 | 92.1 | 91.2 | 41.3 |
| isoCirc HEK293 SRR10612051 | 31.6 | 68.0 | 87.7 | 89.5 | 91.4 | 40.4 |
| isoCirc HEK293 SRR10612052 | 28.5 | 65.4 | 85.0 | 89.9 | 90.9 | 37.8 |
| isoCirc HEK293 SRR10612053 | 37.6 | 73.8 | 89.0 | 92.4 | 91.3 | 47.5 |
| isoCirc HEK293 SRR10612054 | 36.8 | 72.9 | 88.5 | 90.7 | 91.4 | 46.6 |
| isoCirc HEK293 SRR10612055 | 40.9 | 78.6 | 91.1 | 93.0 | 91.9 | 50.0 |
| isoCirc HEK293 all | 18.2 | 51.9 | 77.3 | 85.9 | 94.5 | 34.8 |
| circFL-seq HEK293 | 63.5 | 78.2 | 84.0 | 89.5 | 97.2 | 75.3 |
Additional files
-
Supplementary file 1
Data summary of circFL-seq library.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp1-v2.docx
-
Supplementary file 2
Summary of alternative splicing events of circRNAs detected by circFL-seq.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp2-v2.docx
-
Supplementary file 3
Data summary of RNA-seq library.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp3-v2.docx
-
Supplementary file 4
Comparison of isoCirc and circFL-seq for circRNA detection in the HEK293 cell line.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp4-v2.docx
-
Supplementary file 5
Computational analysis of circFL-seq and CIRI-long.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp5-v2.docx
-
Supplementary file 6
Comparisons between circFL-seq, CIRI-long, and isoCirc.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp6-v2.docx
-
Supplementary file 7
Sequences of hybrid probes for rRNA degradation.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp7-v2.xlsx
-
Supplementary file 8
Summary of performance of strand classifier.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp8-v2.docx
-
Supplementary file 9
Primers to validate rolling circles of circRNAs.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp9-v2.xlsx
-
Supplementary file 10
Primers to validate alternative splicing of circRNAs.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp10-v2.xlsx
-
Supplementary file 11
Primers to validate the expression levels of circRNA BSJs by RT-qPCR.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp11-v2.xlsx
-
Supplementary file 12
Primers to validate the expression levels of circRNA isoforms by RT-qPCR.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp12-v2.xlsx
-
Supplementary file 13
Primers to validate full-length sequence of f-circRNA.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp13-v2.xlsx
-
Supplementary file 14
Primers to validate the expression levels of f-circRNA junctions by RT-qPCR.
- https://cdn.elifesciences.org/articles/69457/elife-69457-supp14-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69457/elife-69457-transrepform1-v2.docx