Identification of bipotent progenitors that give rise to myogenic and connective tissues in mouse
Figures
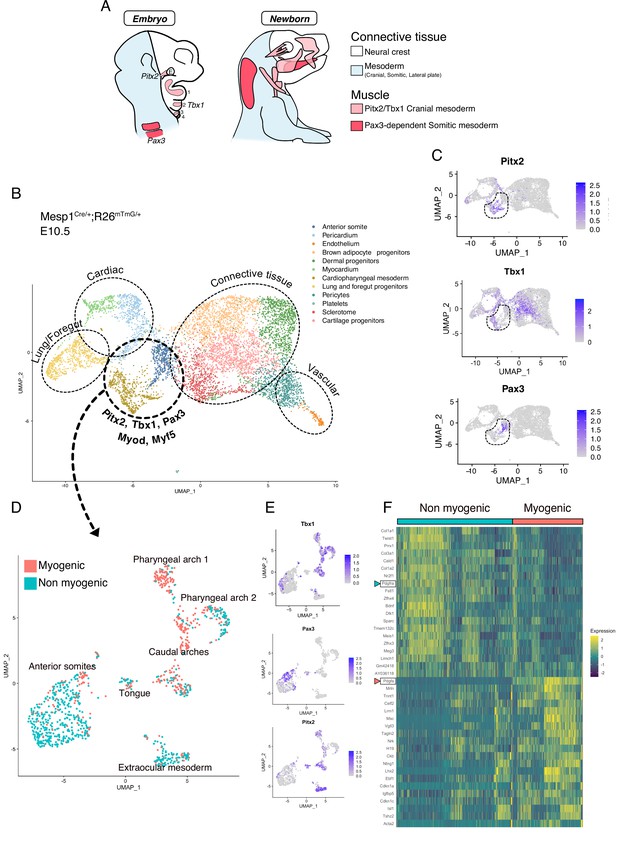
scRNAseq reveals non-myogenic populations of cranial mesoderm lineages.
(A) Scheme of connective tissue origin in the head and known mesodermal upstream regulators. E: Eye, 1–4: Pharyngeal arches 1–4. (B–F) scRNAseq analysis on Mesp1Cre/+; Rosa26mTmG/+ embryos at E10.5 (2 datasets of 2 embryos were aggregated to generate this data, see methods). (B) UMAP of Mesp1Cre/+; Rosa26mTmG/+ E10.5 scRNAseq with main cell types highlighted. The clusters ‘Anterior somite’ and ‘Cardiopharyngeal mesoderm’ were subsetted for further analysis below. (C) UMAP expression plots of Pitx2 (EOM), Tbx1 (cranial mesoderm except EOM) and Pax3 (somitic mesoderm), indicating the clusters of progenitors that were selected. (D) UMAP of progenitor subset annotated as myogenic and non-myogenic based on expression patterns found in E and F. (E) UMAP expression plots of Pitx2, Tbx1 and Pax3 in the Mesp1Cre/+; Rosa26mTmG/+ E10.5 subset. (F) Heatmap of top 20 markers of myogenic versus non-myogenic clusters Mesp1Cre/+; Rosa26mTmG/+ E10.5 subset. Pdgfra/Pdgfa genes are highlighted.
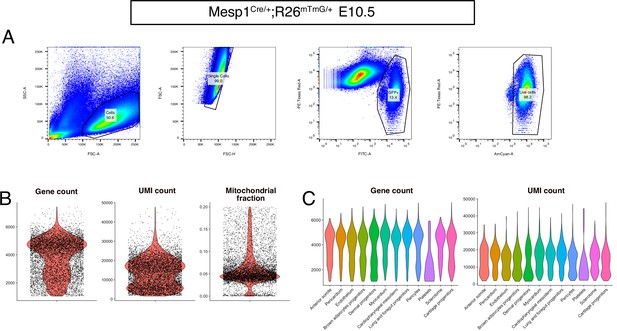
FACS strategy and preprocessing metrics of the Mesp1-derived E10.5 dataset.
(A) Gating strategy used to isolate by FACS Mesp1Cre/+; Rosa26mTmG/+ cells. The FITC channel was used to identify GFP+ cells. The AmCyan channel was used to identify the Calcein Blue+ live cells. The PE-Texas Red channel was used to discard mTomato+ cells and Propidium Iodide+ cells. The percentage of cells captured by each gate is displayed on each plot. (B) Violin plots of gene count, UMI count and mitochondrial fraction for overall dataset. (C) Violin plots of gene count and UMI count by cluster (n = 2 pooled datasets).
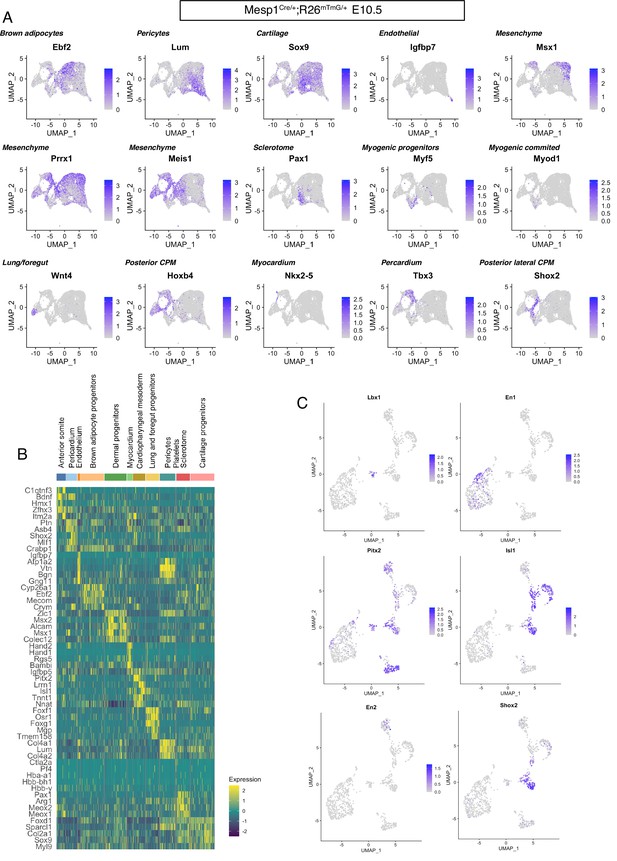
Genetic markers defining anterior mesodermal tissues.
(A) Mesp1Cre/+;Rosa26mTmG/+ E10.5 UMAP expression plots of markers of various mesodermal lineages. (B) Heatmap of top 5 markers of each cluster of Mesp1Cre/+;Rosa26mTmG/+ E10.5. (C) UMAP expression plot of the Mesp1Cre/+;Rosa26mTmG/+ E10.5 subset. En2: marker of pharyngeal arch 1 (Knight et al., 2008), En1: marker of epaxial somitic progenitors (Spörle, 2001), Lbx1: marker of tongue progenitors (Gross et al., 2000), Isl1: marker of cardiopharyngeal mesoderm of pharyngeal arch 2–6 (Nathan et al., 2008), Shox2: marker of caudal cardiopharyngeal mesoderm (Wang et al., 2020), Pitx2: marker of the extraocular region (Zacharias et al., 2011) (n = 2 pooled datasets).
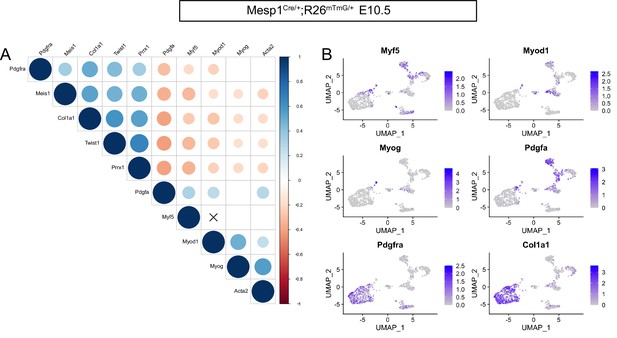
Complementary Pdgf signaling defines myogenic and non-myogenic mesodermal cells.
(A) Pearson correlation plot of myogenic (Pdgfa, Myf5, Myod1, Myog, Acta2) and non-myogenic (Pdgfra, Prrx1, Meis1, Twist1, Osr1, Col1a1) genes in the Mesp1Cre/+;Rosa26mTmG/+ E10.5 subset. The size of the dots is inversely proportional to their p-value. A cross indicates a p-value > 0.05. The color of the dots indicates the strength of the positive (blue) or negative (red) correlation. (B) Expression patterns of Myf5, Myod, Myog, Pdgfa, Pdgfra, and Col1a1 in the Mesp1Cre/+;Rosa26mTmG/+ E10.5 subset dataset. Note that Myf5+ cells were overwhelmingly Pdgfra- and Myf5+/Pdgfra+ cells represent 8% of all cells (i.e. expressing at least one transcript of both genes). Pdgfra+ cells represent 40% of all cells (n = 2 pooled datasets).
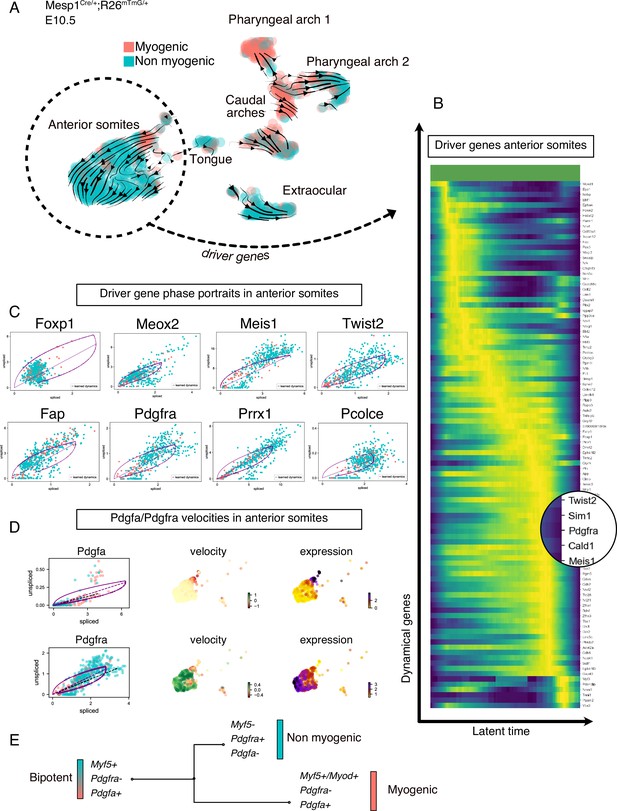
Transcriptomic dynamics reveal a myogenic to non-myogenic transition in anterior somite progenitors.
(A) Velocity UMAP plots of Mesp1Cre/+; Rosa26mTmG/+ embryos at E10.5 displaying myogenic and non-myogenic clusters. Arrows represent the lineage progression based on RNA velocity (relative abundance of unspliced and spliced transcripts). (B) Heatmap of driver genes accounting for anterior somite velocity, highlighting Pdgfra. Driver genes are genes that are transcriptomically active in a given cluster. (C) Phase portraits of few selected driver genes in the anterior somites: Foxp1, Meox2, Meis1, Twist2, Fap, Pdgfra, Prrx1, and Pcolce. Y-axis represents the amount of unspliced transcript per cell; X-axis represents the number of spliced transcripts per cell. A high fraction of unspliced variants indicates an active transcription of the locus, while the inverse indicates inactive/repressed transcription. Dynamics of transcription were inferred at a gene- and cluster-specific level (see Methods). (D) Phase portraits, RNA velocity and expression plots of Pdgfa and Pdgfra showing splicing dynamics of these two genes. (E) Working model of myogenic and non-myogenic fate decisions from a common bipotent progenitor in anterior somites.
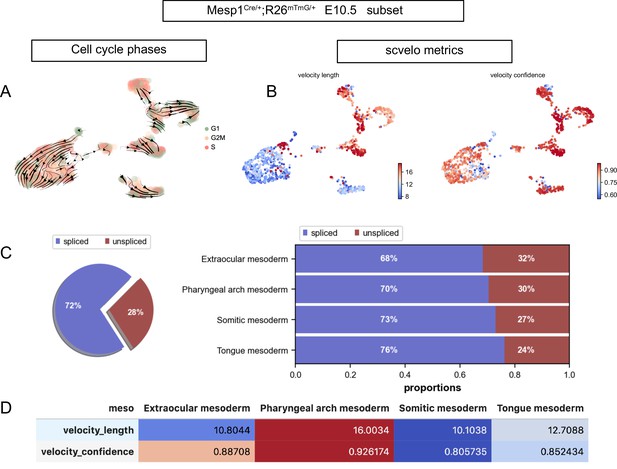
Cell cycle phases and RNA velocity metrics of the Mesp1-derived E10.5 dataset.
(A) UMAP of Mesp1Cre/+;Rosa26mTmG/+ E10.5 subset with overlaid velocity and cell cycle phase. (B–D) Quality control metrics of scvelo, including velocity length, velocity confidence and spliced/unspliced abundance in the overall dataset and by cluster (n = 2 pooled datasets).
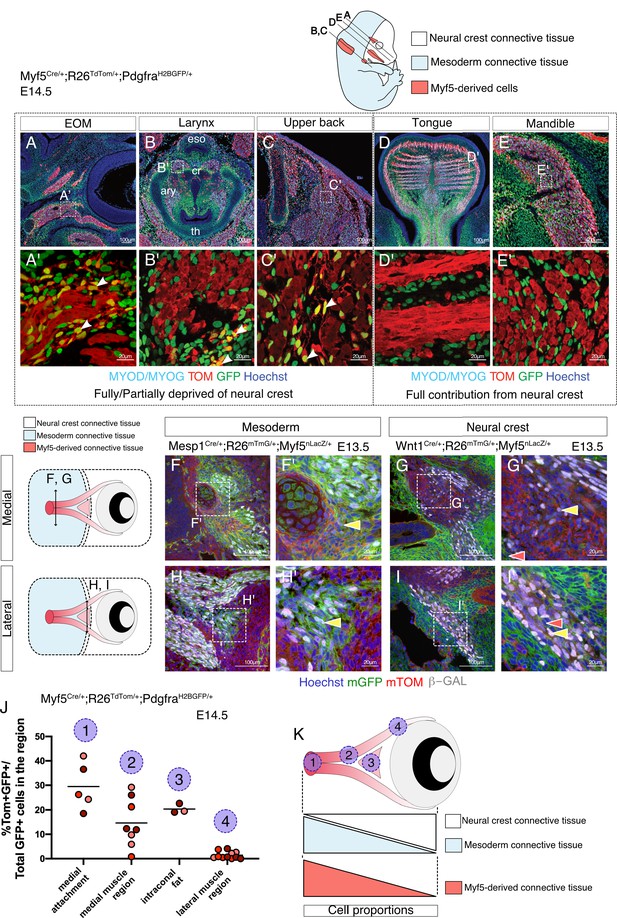
Myf5-derived mesodermal connective tissue partially compensates for the lack of neural crest.
(A-E') Transverse sections of an E14.5 Myf5Cre/+; Rosa26TdTomato/+; PdgfraH2BGFP/+ embryo immunostained for Myod/Myog. White arrowheads indicate cells double-positive GFP/TOM and negative for Myod/Myog (n = 3 embryos). (F-I') Transverse cryosections of the EOM at E13.5 of Wnt1Cre/+; Rosa26mTmG/+; Myf5nlacZ/+ (G,I) and Mesp1Cre/+; Rosa26mTmG/+; Myf5nlacZ/+ (F,H) immunostained for β-gal, at the level of the medial attachment (F,G) and lateral muscle masses (H,I). Yellow arrowheads indicate Myf5-expressing cells in the context of mesodermal and neural crest lineages. Note that Myf5-expressing cells are mGFP+ in the Mesp1 lineage and mGFP- in the Wnt1 lineage. Red arrowheads indicate neural-crest cells that are excluded from the Myf5 lineage (n = 2 embryos for each). (J) Scatter plots of the proportion of double positive cells in E14.5 Myf5Cre/+; Rosa26TdTomato/+; PdgfraH2BGFP/+ embryos in various regions throughout the EOM (the line is the mean, each dot is a tissue section, each color is a different embryo, n = 3 embryos). (K) Scheme highlighting the quantified regions in (J) and summarising the contribution of each population to periocular connective tissues. TOM: TdTOMATO.
-
Figure 3—source data 1
Excel table summarizing the quantification displayed on Figure 3J.
- https://cdn.elifesciences.org/articles/70235/elife-70235-fig3-data1-v3.xlsx
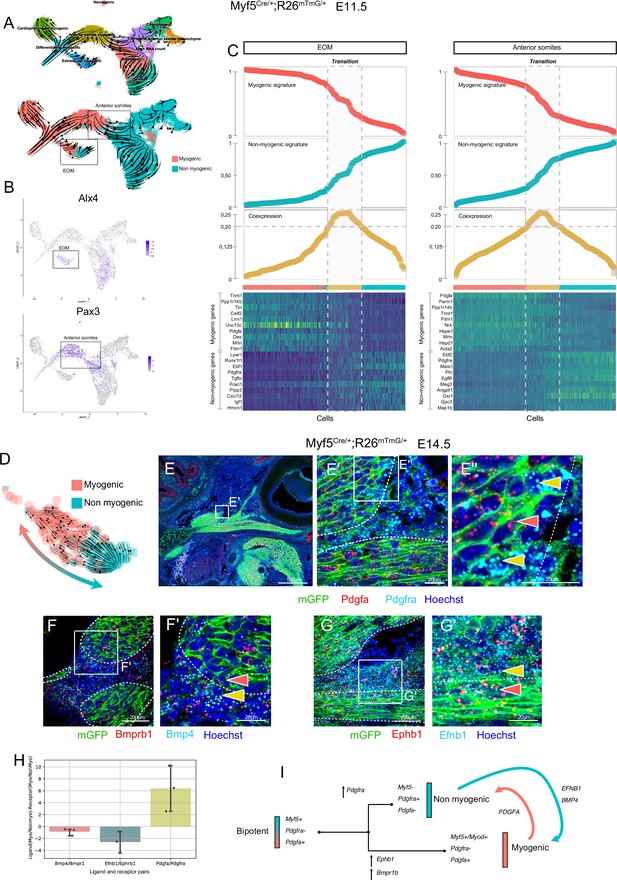
Maintenance of signaling cues between Myf5-derived myogenic and non-myogenic cells in EOM.
(A–D) scRNAseq analysis of the Myf5Cre/+; Rosa26mTmG/+ E11.5 dataset (2 datasets of 2 embryos were aggregated to generate this data, see Materials and methods). (A) UMAPs of Myf5Cre/+; Rosa26mTmG/+ E11.5 RNA velocity trajectories. (B) Expression plots of Alx4 and Pax3, highlighting EOM and Anterior somite clusters, respectively. (C) Plots of Myogenic and Non-myogenic signatures, Coexpression score and heatmaps of top markers, highlighting the transition population in EOM and anterior somites. Cells are ordered based on their non-myogenic signature score (increasing). The coexpression score is the product of the myogenic and non-myogenic signatures. Cells presenting a coexpression score higher than 0.20 are highlighted in yellow. These cells represent the transition between the myogenic and non-myogenic fates. (D) UMAP of the EOM subset revealing the bipartite fate of Myf5-expressing cells. (E-G’) RNAscope on Myf5Cre/+; Rosa26mTmG/+ E14.5 tissue sections with Pdgfra (cyan) and Pdgfa (red) probes (E-E’’), Bmprb1 (red) and Bmp4 (cyan) probes (F-F’) and Ephb1 (red) and Efnb1 (cyan) probes (G-G’). Myf5-derived cells are labelled by membrane GFP staining (n = 3 embryos each). Red and yellow arrowheads indicate Myf5-derived myogenic and non-myogenic cells respectively. The dotted lines highlight the boundary of the muscle masses. (H) Quantification of the Ligand-Receptor scores for each pair (see Materials and methods). Note that these ratios are negative in the case of Bmp and Eph (signaling from non-myogenic to myogenic) but positive for Pdgf (signaling from myogenic to non-myogenic). (G) Model of myogenic and non-myogenic cell communication following bifurcation from a bipotent cell.
-
Figure 4—source data 1
Excel table summarizing the quantification displayed on Figure 4H.
- https://cdn.elifesciences.org/articles/70235/elife-70235-fig4-data1-v3.xlsx
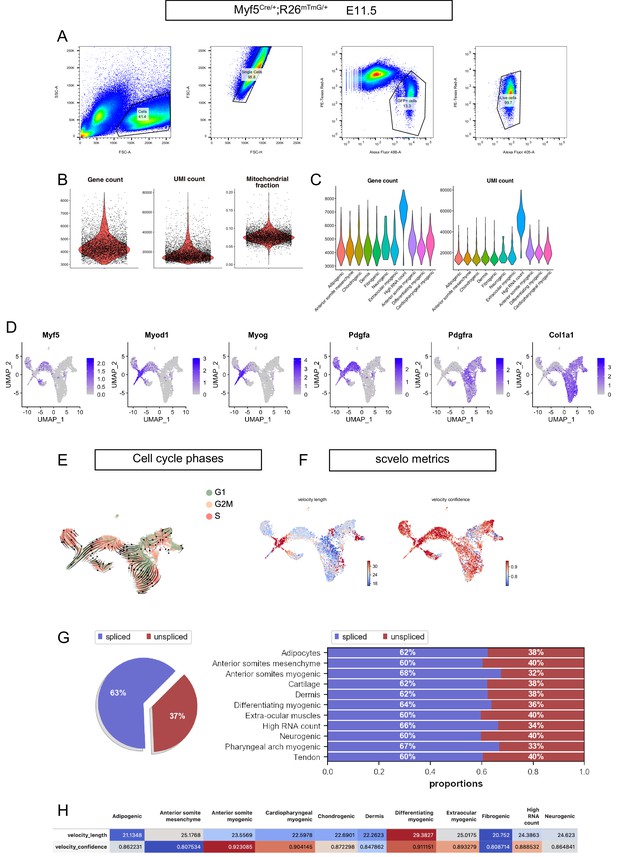
FACS strategy, preprocessing metrics, expression patterns, and RNA velocity metrics of the Myf5-derived E11.5 dataset.
(A) Gating strategy used to isolate by FACS Myf5Cre/+; Rosa26mTmG/+ cells. The Alexa Fluor 488 channel was used to identify GFP+ cells. The Alexa Fluor 405 channel was used to identify the Calcein Blue+ live cells. The PE-Texas Red channel was used to discard mTomato+ cells (non recombined) and Propidium Iodide+ cells. The percentage of cells captured by each gate is displayed on each plot. (B) Violin plots of gene count, UMI count and mitochondrial fraction for overall dataset. (C) Violin plots of gene count and UMI count by cluster. (D) Expression patterns of Myf5, Myod, Myog, Pdgfa, Pdgfra, and Col1a1 in the Myf5Cre/+; Rosa26mTmG/+ E11.5 dataset. Note that Myf5+ cells were overwhelmingly Pdgfra- and Myf5+/Pdgfra+ cells represent 5.5% of all cells (i.e. expressing at least one transcript of both genes). Pdgfra+ cells represent 56% of all cells. (E) UMAP of Myf5Cre/+; Rosa26mTmG/+ E11.5 with overlaid velocity and cell cycle phase. (F–H) Quality control metrics of scvelo, including velocity length, velocity confidence and spliced/unspliced abundance in overall dataset and by cluster (n = 2 pooled datasets).
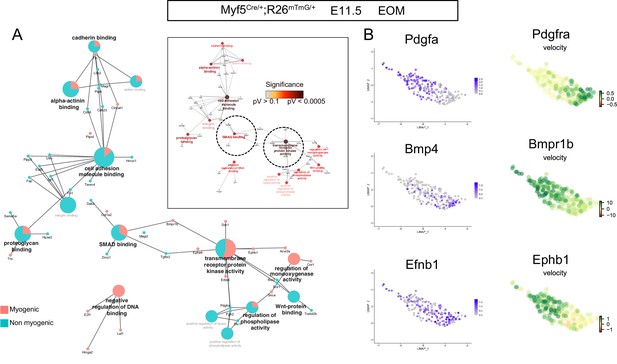
Kinase signaling complementarity in the EOM at E11.5.
(A) GO Molecular Function network of top 100 driver genes of the Myf5Cre/+; Rosa26mTmG/+ E11.5 EOM dataset (see Table 1), including relative contribution of each cluster (myogenic and non-myogenic) to the term and significance levels. Insert show the significance of each term. (B) UMAPs of the Myf5Cre/+; Rosa26mTmG/+ E11.5 EOM dataset showing the expression of kinase ligands (left side) and the velocity of their corresponding receptors (right side). Note the complementary patterns of the L/R pairs (n = 2 pooled datasets).
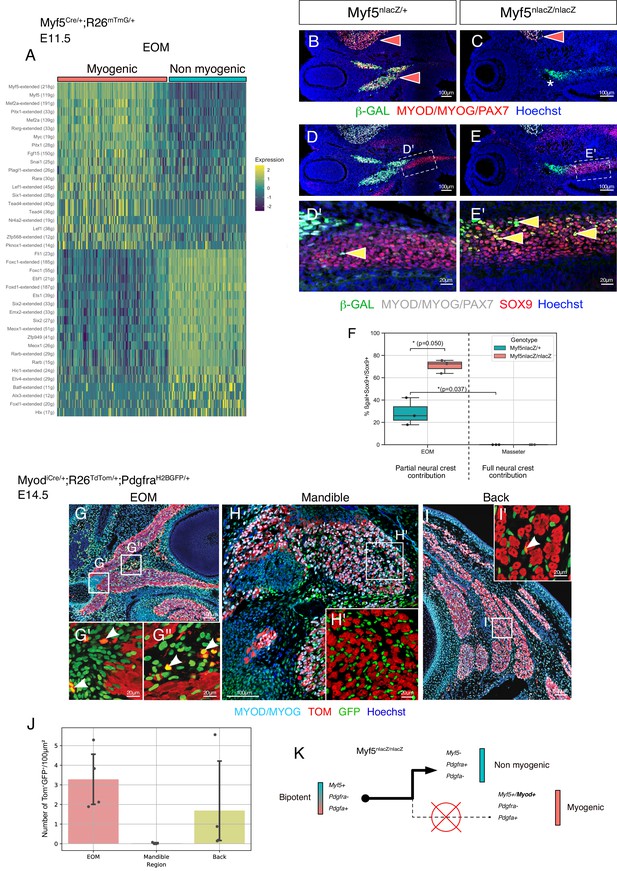
Disruption of Myf5 increases the connective tissue output from bipotent cells.
(A) Heatmap of top regulons (transcription factor and associated targets) of the EOM subset of the Myf5Cre/+; Rosa26mTmG/+ E11.5 dataset. The suffix ‘_extended’ indicates that the regulon includes motifs that have been linked to the TF by lower confidence annotations, for instance, inferred by motif similarity. Number in brackets indicates number of genes comprising the regulon (n = 2 pooled datasets). (B–C) Transverse sections of Myf5nlacZ/+ (B), and Myf5nlacZ/nlacZ (C) embryos in the EOM region at E12.5 immunostained for β-gal (green), and Myod/Myog/Pax7 (red). Red arrowheads indicate β-gal/ Myod/Myog/Pax7 double positive cells in control EOM/Masseter and in mutant Masseter. Asterisk highlights the lack of myogenic progenitors in the EOM region of the mutant embryo, indicated by the absence of Myod/Myog/Pax7 staining. (D-E’) Transverse sections of Myf5nlacZ/+ (D-D'), and Myf5nlacZ/nlacZ (E-E') in the EOM region at E12.5 immunostained for β-gal (green), Sox9 (red), and Myod/Myog/Pax7 (gray). Yellow arrowheads indicate β-gal/Sox9 double positive cells and show an expansion of this cell population in the mutant. (F) Quantification of proportion of β-gal+;Sox9+ double positive cells in the total Sox9+ population of the EOM and Masseter muscles. Each dot is a different sample, the center line of the boxplot is the median value. (n = 3 embryos, p-values were calculated using a two-sided Mann-Whitney U test). (G-I’) Transverse sections of MyodiCre/+; Rosa26TdTomato/+; PdgfraH2BGFP/+ embryos at E14.5 immunostained for Myod/Myog (committed and differentiating myoblasts) in the extraocular (G-G’’), mandibular (H-H’), and back muscles (I-I’). White arrowhead indicates double positive cells (GFP+ TOM+). (J) Quantification of double positive cells (GFP+ TOM+) in EOM, mandibular muscles and back muscles per 100 μm2 area on MyodiCre/+; Rosa26TdTomato/+; PdgfraH2BGFP/+ sections shown in E-G (n = 4 embryos). (K) Model of lineage progression from bipotent cells in a Myf5 null background.
-
Figure 5—source data 1
Excel table summarizing the quantification displayed on Figure 5F.
- https://cdn.elifesciences.org/articles/70235/elife-70235-fig5-data1-v3.xlsx
-
Figure 5—source data 2
Excel table summarizing the quantification displayed on Figure 5J.
- https://cdn.elifesciences.org/articles/70235/elife-70235-fig5-data2-v3.xlsx
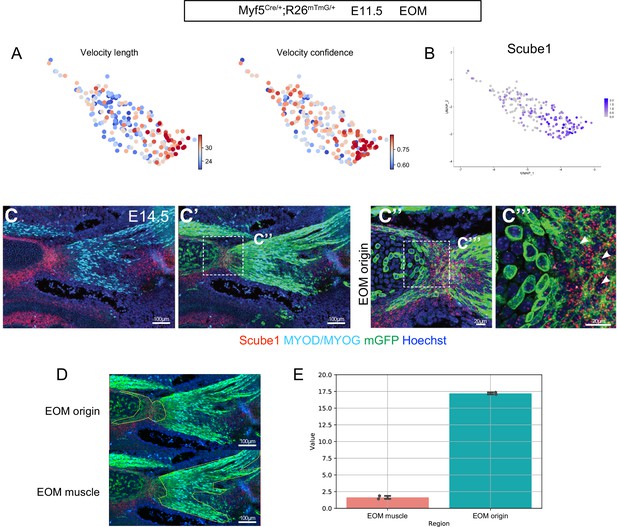
The vascular marker Scube1 is expressed in Myf5-derived non-myogenic cells in the EOM.
(A) UMAP of Myf5Cre/+; Rosa26mTmG/+ E11.5 EOM dataset illustrating velocity confidence and velocity length. Higher confidence is found on both ends of the EOM cluster. (B) Expression pattern of Scube1 in the EOM subset (mostly in the non-myogenic compartment). (C-C’’) Combined RNAscope for Scube1 and Myod/Myog/GFP immunostaining on transverse sections of Myf5Cre/+; Rosa26mTmG/+ at E14.5. (D) Model of the EOM muscle vs EOM origin compartmentalization used for quantification in (E). (E) Quantification of Scube1 signal in each compartment (n = 2 embryos).
-
Figure 5—figure supplement 1—source data 1
Excel table summarizing the quantification displayed on Figure 5—figure supplement 1E.
- https://cdn.elifesciences.org/articles/70235/elife-70235-fig5-figsupp1-data1-v3.xlsx
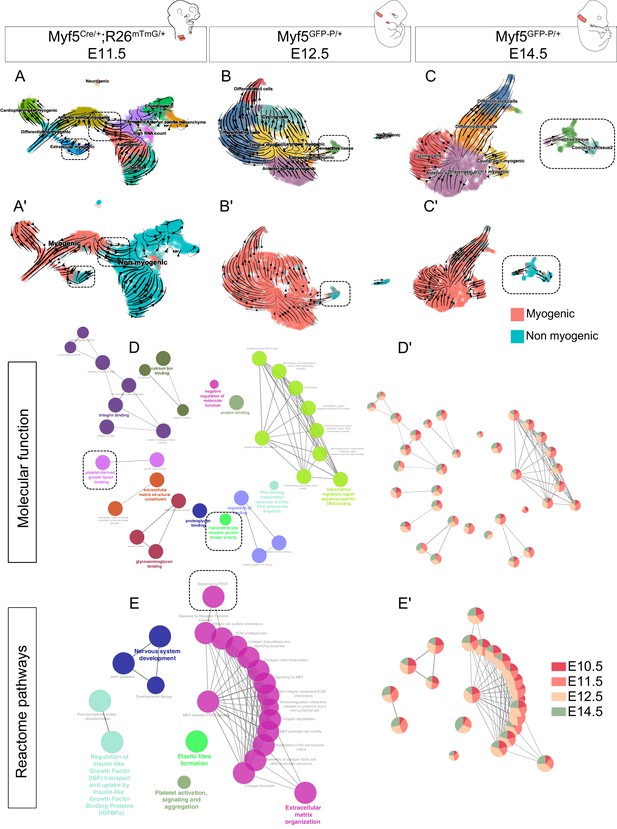
Myf5-derived non-myogenic cells are generated continuously up to fetal stages.
(A-C') RNA velocity plots of Myf5Cre/+; Rosa26mTmG/+ E11.5, Myf5GFP-P/+ E12.5 and Myf5GFP-P/+ E14.5 datasets (n = 2 pooled datasets, n = 1 embryo and n = 1 embryo, respectively) displaying cell-type annotation (A–C) and myogenic and non-myogenic clustering (A’-C’). The dotted boxes highlight the transitions to non-myogenic clusters in each dataset. (D–E) Gene ontology network of GO Molecular Function and Reactome pathway performed on combined top 100 markers using Cluego. These terms were generated using the sum of all differentially expressed genes of the non-myogenic clusters across all datasets (see Materials and methods). (D’-E’) Relative contribution of each stage to term node represented as piecharts (i.e. the proportion of genes underlying this term coming from that stage). Dotted boxes highlight the shared tyrosine kinase and PDGF signaling pathways.
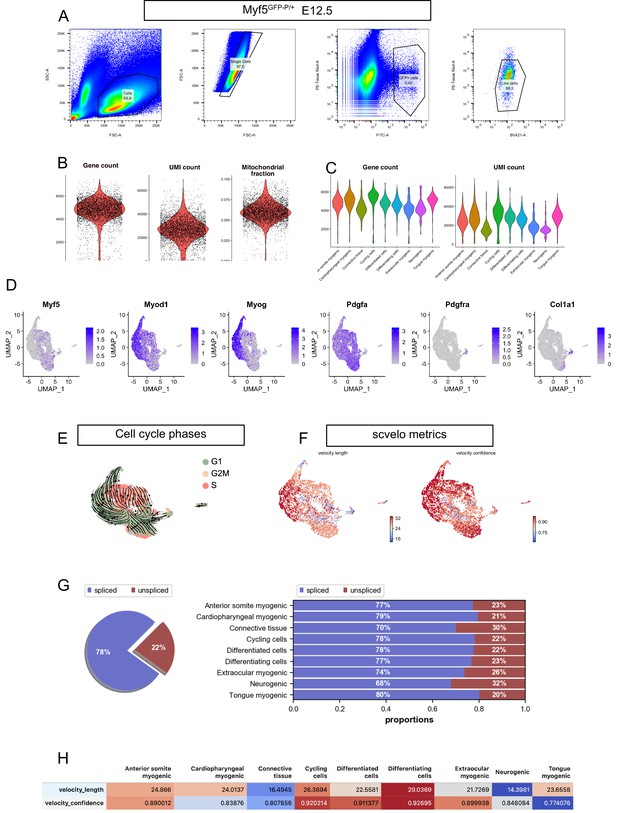
FACS strategy, preprocessing metrics, expression patterns, and RNA velocity metrics in the Myf5GFP-P/+E12.5 dataset.
(A) Gating strategy used to isolate by FACS Myf5GFP-P/+ cells. The FITC channel was used to identify GFP+ cells. The BV421 was used to identify the Calcein Blue+ live cells. The PE-Texas Red channel was used to discard Propidium Iodide+ cells. The percentage of cells captured by each gate is displayed on each plot. (B) Violin plots of gene count, UMI count and mitochondrial fraction for overall dataset. (C) Violin plots of gene count and UMI count by cluster. (D) Expression patterns of Myf5, Myod, Myog, Pdgfa, Pdgfra, and Col1a1 in the Myf5GFP-P/+ E12.5 dataset. Note that Myf5+ cells were overwhelmingly Pdgfra- and Myf5+/Pdgfra+ cells represent 0.5% of all cells (i.e. expressing at least one transcript of both genes). Pdgfra+ cells represent 3% of all cells. (E) UMAP of Myf5GFP-P/+ E12.5 with overlaid velocity and cell cycle phase. (F–H) Quality control metrics of scvelo, including velocity length, velocity confidence and spliced/unspliced abundance in overall dataset and by cluster (n = 1 embryo).
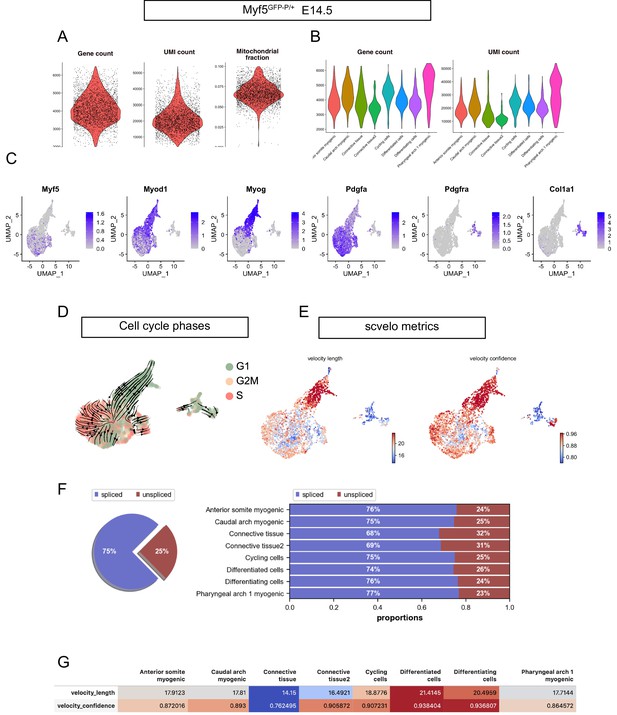
FACS strategy, preprocessing metrics, expression patterns and RNA velocity metrics in the Myf5GFP-P/+ E14.5 dataset.
(A) Violin plots of gene count, UMI count and mitochondrial fraction for overall dataset. (B) Violin plots of gene count and UMI count by cluster. (C) Expression patterns of Myf5, Myod, Myog, Pdgfa, Pdgfra, and Col1a1 in the Myf5GFP-P/+ E14.5 dataset. Note that Myf5+ cells were overwhelmingly Pdgfra- and Myf5+/Pdgfra+ cells represent 0.15% of all cells (i.e. expressing at least one transcript of both genes). Pdgfra+ cells represent 7% of all cells (n = 1 embryo). (D) UMAP of Myf5GFP-P/+ E14.5 with overlaid velocity and cell cycle phase (n = 1 embryo). (E–G) Quality control metrics of scvelo, including velocity length, velocity confidence and spliced/unspliced abundance in overall dataset and by cluster (n = 1 embryo).
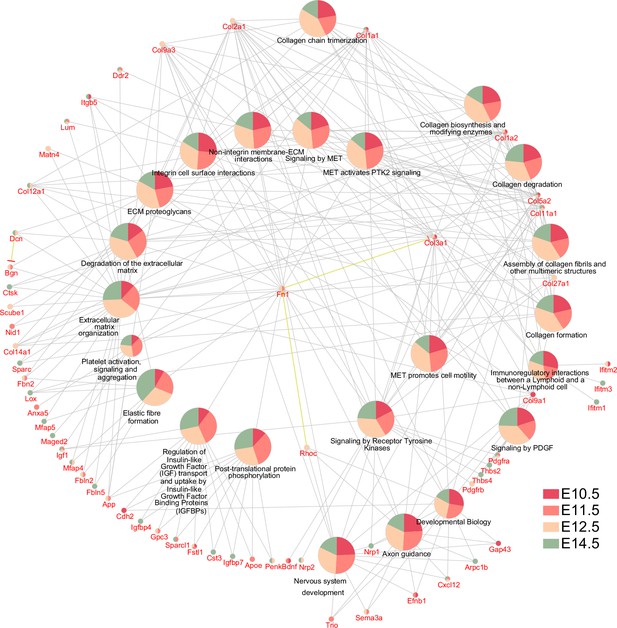
Non-myogenic Myf5-derived cells display a similar gene ontology.
Gene ontology analysis for Reactome pathways, including genes underlying each term, and their representation in each dataset generated using Cluego based on top 100 differentially expressed genes of the non-myogenic clusters (see Materials and methods, E10.5: n = 2 pooled datasets, E11.5: n = 2 pooled datasets, E12.5: n = 1 embryo and E14.5: n = 1 embryo).
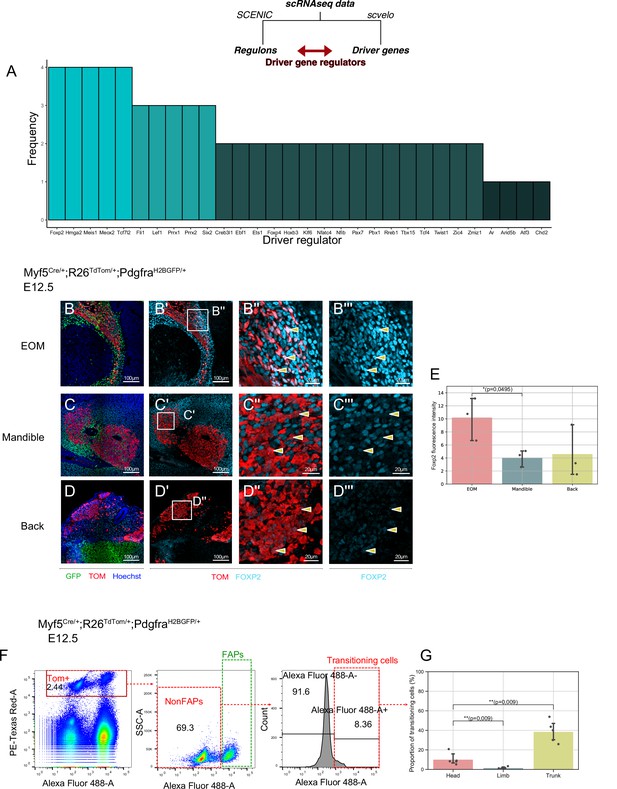
A shared program involving Forkhead-box transcription factors supports non-myogenic fate transition at various stages and anatomical locations.
(A) Barplot displaying frequency of appearance of most predominant transcription factors as driver regulators (4 = present in all four datasets as driver regulon, 1 = present in a single dataset). (B-D’’) Transverse sections of an E12.5 Myf5Cre/+; Rosa26TdTomato/+; PdgfraH2BGFP/+ embryo immunostained for Foxp2 at the level of the EOM (B-B’’), Mandibular muscles (C-C’’), and Back muscles (D-D’’). Yellow arrowheads indicated the double positive cells to better appreciate Foxp2 intensity in Myf5-derived cells. (E) Quantification of Foxp2 signal intensity in TOM+ (Myf5-derived) cells in each muscle (n = 3 embryos). Statistical test performed: Mann-Whitney U test. (F) FACS plots of dissected E12.5 Myf5Cre/+; Rosa26TdTomato/+; PdgfraH2BGFP/+ embryos (head region here) highlighting the Myf5-derived GFP- TOM+ population transitioning to the GFP+ TOM+ population. Each plot was generated on the population gated in the previous one (‘Singlets’, ‘TOM+’ and ‘NonFaps’). FAPS:Fibroadipogenic progenitors, a denomination for resident Pdgfra+ cells. (G) Quantification of the transitioning population in Head, Limb and Trunk. Proportion of transitioning cells is calculated as the number of Alexa488+/Total cell number in the ‘NonFAPs’ gate. Note that the Head region is mostly populated by muscles embedded in neural crest (n = 5 embryos). TOM: TdTOMATO.
-
Figure 7—source data 1
Excel table summarizing the quantification displayed on Figure 7E.
- https://cdn.elifesciences.org/articles/70235/elife-70235-fig7-data1-v3.xlsx
-
Figure 7—source data 2
Excel table summarizing the quantification displayed on Figure 7G.
- https://cdn.elifesciences.org/articles/70235/elife-70235-fig7-data2-v3.xlsx
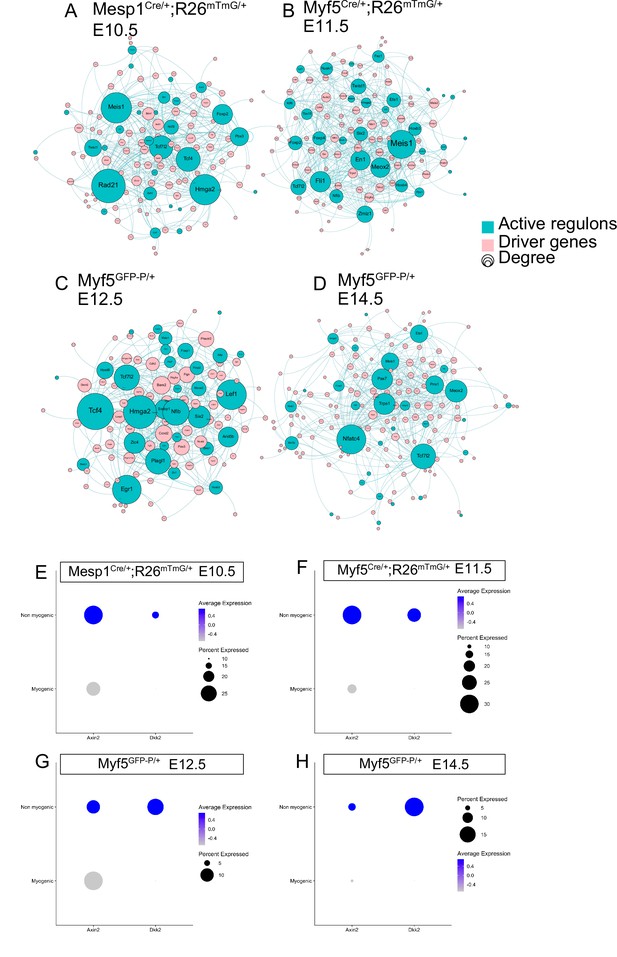
Wnt/β-cat positive feedback loop may promote non-myogenic cell fate.
(A–D) Driver genes and regulatory networks (regulons) were produced for each stage independently, and a stage-specific network of active transcription factor and associated driver gene targets was built (n = 2 pooled datasets, n = 2 pooled datasets, n = 1 embryo and n = 1 embry, respectively). The size of nodes corresponds to the number of edges (connections) they have, i.e. the number of driver genes the transcription factor regulates. (E–H) Dotplot of the expression levels and percent of Axin2 and Dkk2 in the myogenic and the non-myogenic portions of all four datasets.
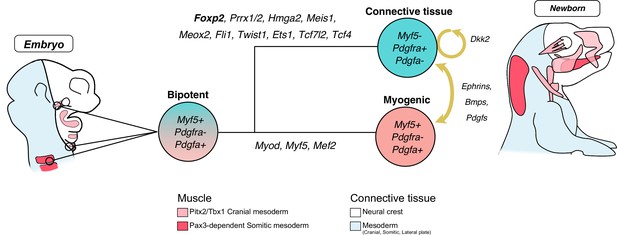
Model of Myf5+ bipotent progenitors giving rise to muscle and associated connective tissues.
Model for bipotent Myf5+/Pdgfa+ progenitors giving rise to myogenic and non-myogenic cells; discrete parts of the head deprived of neural crest are indicated. Upon activation of a set of transcription factors including Prrx1/2, Foxp2, Hmga2, Meis1, Meox2, Fli1, Twist1, Ets1, Tcf7l2, and Tcf4, a fibrogenic fate is acquired. A molecular dialogue is initiated at the branchpoint including extracellular matrix components and kinase signalling such as Pdgf, Ephrins, and Bmps. The non-myogenic fate may be maintained cell-autonomously by a canonical Wnt-positive feedback loop.
Tables
Driver genes underlying cell fate decisions in each dataset.
| E10.5 Anterior somites | E11.5 EOM Myogenic | E11.5 EOM Non-myogenic | E12.5 Non-myogenic | E14.5 Non-myogenic |
|---|---|---|---|---|
| Tshz2 | Ccdc141 | Zfpm2 | Mgat4c | Dnm1 |
| Eya1 | Mcm6 | Plxna4 | Cenpv | Pid1 |
| C1qtnf3 | Dync1i1 | Col23a1 | C130073E24Rik | Nrp2 |
| Meis2 | Tpm2 | Edil3 | Tbx3os1 | Ntrk3 |
| Limch1 | Celf2 | Map2 | E330013P04Rik | Tmem132c |
| Moxd1 | Sox6 | Rora | Stk26 | Egflam |
| Epha4 | Tnc | Sema5a | Edil3 | Gpr153 |
| Pitx2 | Magi3 | Colec12 | Fdft1 | Efemp1 |
| Parm1 | Sh3glb1 | Smoc1 | Lima1 | Adamts2 |
| Hpse2 | Parm1 | Ptprt | Trim59 | Brinp1 |
| Lrrn1 | Ephb1 | Ror1 | Meg3 | Vegfc |
| Dmrt2 | Bmpr1b | Dock5 | Gins3 | Twist2 |
| Myl3 | Hells | Map1b | Tpm2 | Itgb5 |
| Fap | Pdgfc | Fn1 | Cdh6 | Gria1 |
| Hs6st2 | Ptprd | Limch1 | Csmd3 | Sned1 |
| Ddr2 | Cnr1 | Tenm4 | Tceal5 | Sorcs3 |
| Cald1 | Sema3d | Rbms3 | Pclaf | Ebf2 |
| Prrx1 | Clcn5 | Srgap3 | Tspan9 | Fam19a1 |
| Magi3 | Chd7 | Tmem132c | Eps8 | Trabd2b |
| Ntn1 | Col25a1 | Sdc2 | Lmna | Plxdc2 |
| Zfhx3 | Reep1 | Add3 | Dmrt2 | Sh3gl3 |
| Meis1 | Ctnnal1 | Pdgfra | Cpeb4 | Luzp2 |
| Tnni1 | Tpm1 | Gmds | Hpgd | Pdzd2 |
| Crym | Zim1 | St6galnac3 | Rcsd1 | Sema3e |
| Ebf1 | Lmx1a | Epb41l3 | Pdgfra | Rims1 |
| Nr2f1 | Neb | Pde3a | Plac1 | Epha3 |
| Ntng1 | Atad2 | Tox | Palmd | Cyp7b1 |
| Pgm5 | Dapk2 | Smarca2 | Gucy1a1 | Gem |
| Cdh6 | Prox1 | Ctdspl | Wif1 | Ldb2 |
| Foxp1 | Lsamp | Magi2 | Naalad2 | Scube1 |
| Celf2 | Ttn | Dpysl3 | Smoc2 | Pdgfra |
| Tbx1 | Pls3 | Fgfr2 | Rassf4 | Pde1a |
| Bdnf | Slf2 | Ldb2 | Pttg1 | Nde1 |
| Colec12 | Vat1l | Igf1 | Josd2 | Enpp2 |
| Eya4 | E2f1 | Elk3 | Plxna4 | Fam107b |
| Sobp | Epb41l2 | Zmiz1 | Eya2 | Stxbp6 |
| Peg3 | Gm28653 | Dlc1 | Nrsn1 | Rerg |
| Pdgfra | Lrrn1 | Nhs | Fign | Prex2 |
| Nrk | Mef2c | Cdkn1c | Inppl1 | Man1a |
| Ptn | St8sia2 | Plpp3 | Rnf152 | Tmem45a |
| Daam1 | Tshz1 | Ebf1 | Lasp1 | Sh3bp4 |
| Dlk1 | Wee1 | Sorbs2 | Mrln | Mcc |
| Unc5c | Slc24a3 | Baz1a | Cdt1 | Ncald |
| Lpar1 | Ncoa1 | Fat4 | Notch3 | Kdelr2 |
| Syne2 | Dek | Golgb1 | Pax3 | Pcdh19 |
| Nkd2 | Kdm5b | Hpse2 | Egfr | Gas7 |
| Brinp1 | Unc13c | Samd4 | Dbf4 | Cpt1c |
| Zfhx4 | Ddr1 | Itga9 | Bcr | Adam22 |
| Nnat | Pip4k2a | Magi1 | Mllt3 | Itgb8 |
| Gxylt2 | Fndc3c1 | Pcdh9 | Nectin1 | Dchs2 |
| Clmp | Rbm24 | Tgfbr2 | Grin3a | Cep350 |
| Ror2 | Rreb1 | Ntf3 | Cbfa2t3 | Oat |
| Nfia | Rragd | Col11a1 | Cdh2 | Rab30 |
| Ebf2 | Acsl3 | Runx1t1 | Anln | Aff2 |
| Ednra | Acvr2a | Tnrc18 | Ccdc6 | Gna14 |
| Fli1 | Zeb1 | Crym | Mcu | Slc29a1 |
| Tspan12 | Rgma | Fap | Fnip2 | Pls3 |
| Ttc28 | Arpp21 | Ppp1r1a | Kcnk13 | Traf3ip1 |
| Nfib | Lef1 | Tes | Sned1 | Rcsd1 |
| Ccdc88c | Nr2f2 | Bicc1 | Nde1 | Lgr4 |
| Col13a1 | Foxo1 | Il1rapl1 | Hipk3 | Zfp9 |
| 2700069I18Rik | Pdzrn4 | Alcam | Arhgap11a | Hs3st5 |
| Pcolce | Hmga2 | 2700069I18Rik | Fam8a1 | Aspn |
| Scn3a | Lurap1l | Dab2 | Kif21a | Nrxn1 |
| Acvr2a | Pkig | Cntln | Mtss1 | Rrm1 |
| Auts2 | Ncl | Clmn | Abcd2 | Igfbp7 |
| Col3a1 | CT025619.1 | Rbms1 | Irx5 | Slc35f3 |
| Gap43 | Erbb4 | Tmem2 | Pacs2 | Kif15 |
| Mrln | Cdk14 | Cdh6 | Nab1 | Slc1a3 |
| Pax3 | Kif21a | Lypd6 | Ccnd2 | Bmp6 |
| Sim1 | Zfp704 | Mmp2 | Bok | Dkk2 |
| Epb41l2 | Nasp | Kif5c | Dok5 | Tspan9 |
| Ppp3ca | Plekha5 | Cadm2 | Ncapg | Ets1 |
| Tnfaip6 | Cap2 | Prkg2 | Rfx8 | Gria3 |
| Tmem132c | Snca | Cped1 | Fhod3 | Sox8 |
| Tmem2 | Epha4 | Dtl | Tk1 | Melk |
| Epb41l3 | Atad5 | Ror2 | Asf1b | Ntm |
| Crybg3 | Cntn3 | Utrn | Tek | Synpo2l |
| Nrxn1 | Cacna2d1 | Foxp1 | Arfgef3 | Hlf |
| Farp1 | Pak3 | L3mbtl3 | Rnf182 | Adamts5 |
| Sulf1 | Megf10 | Cdh23 | Kif14 | Plcb4 |
| Tmtc2 | Tnnt1 | Negr1 | 1810041L15Rik | Cdc25b |
| Pde4dip | Acta2 | Hmcn1 | Rrm2 | Mgat4a |
| Phldb2 | Barx2 | Col26a1 | Fgf5 | Mdfic |
| Plpp3 | Mrln | Fbn2 | Barx2 | Trpc5 |
| Ybx3 | Pgm5 | Ankrd12 | Fli1 | Kif4 |
| Ppm1l | Fmr1 | Lhfp | Jph2 | Plce1 |
| Twist2 | Smc4 | Hs3st3b1 | Dtx4 | Il17rd |
| Nuak1 | Clmp | Adgrl3 | Ncald | Mmp16 |
| Tgfb2 | Alpk2 | Svil | Zic4 | Hhip |
| Sfrp1 | Kctd1 | Mob3b | Dlc1 | Tpx2 |
| Sncaip | Meg3 | Trabd2b | Cdc45 | Ndc80 |
| Tenm3 | Samd5 | Rmst | Gatm | Bub1b |
| Cdh2 | Nrk | Prrx1 | Ssc5d | Hmmr |
| Iqgap2 | Piezo2 | 5330434G04Rik | Phactr2 | Kank4 |
| App | Robo1 | Zfhx3 | Ppp1r14c | Tmeff2 |
| Pgam2 | Col1a2 | Foxp2 | Agl | Nr4a1 |
| Rspo3 | Cntrl | Mpp6 | Tox3 | Aurkb |
| Cdon | Mllt3 | Crispld1 | Aurka | Lrrtm3 |
| Ebf3 | Peg3 | Eya1 | Cdh15 | Cenpq |
Driver regulators of non-myogenic fate in each dataset.
| E10.5 | E11.5 | E12.5 | E14.5 | |
|---|---|---|---|---|
| Foxp2 | (+) | (+) | (+) | (+) |
| Hmga2 | (+) | (+) | (+) | (+) |
| Meis1 | (+) | (+) | (+) | (+) |
| Meox2 | (+) | (+) | (+) | (+) |
| Tcf7l2 | (+) | (+) | (+) | (+) |
| Fli1 | (+) | (+) | (+) | (-) |
| Lef1 | (-) | (+) | (+) | (+) |
| Prrx1 | (+) | (+) | (-) | (+) |
| Prrx2 | (-) | (+) | (+) | (+) |
| Six2 | (+) | (+) | (+) | (-) |
| Creb3l1 | (-) | (+) | (-) | (+) |
| Ebf1 | (+) | (-) | (+) | (-) |
| Ets1 | (-) | (+) | (-) | (+) |
| Foxp4 | (+) | (+) | (-) | (-) |
| Hoxb3 | (-) | (+) | (+) | (-) |
| Klf6 | (-) | (+) | (-) | (+) |
| Nfatc4 | (-) | (+) | (-) | (+) |
| Nfib | (-) | (+) | (+) | (-) |
| Pax7 | (-) | (-) | (+) | (+) |
| Pbx1 | (-) | (+) | (-) | (+) |
| Rreb1 | (-) | (-) | (+) | (+) |
| Tbx15 | (+) | (+) | (-) | (-) |
| Tcf4 | (+) | (-) | (+) | (-) |
| Twist1 | (+) | (+) | (-) | (-) |
| Zic4 | (+) | (-) | (+) | (-) |
| Zmiz1 | (-) | (+) | (+) | (-) |
| Ar | (-) | (-) | (-) | (+) |
| Arid5b | (-) | (-) | (+) | (-) |
| Atf3 | (-) | (-) | (-) | (+) |
| Chd2 | (+) | (-) | (-) | (-) |
-
(+): Present, (-): Absent.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | B6D2F1/JRj | Janvier | ||
| Genetic reagent (M. musculus) | Myf5Cre | PMID:17418413 | MGI:3710099 | Dr. Mario R Capecchi (Institute of Human Genetics, University of Utah, USA) |
| Genetic reagent (M. musculus) | Wnt1Cre | PMID:9843687 | MGI:J:69326 | Pr. Andrew P. McMahon (Keck School of Medicine of the University of Southern California, USA) |
| Genetic reagent (M. musculus) | Mesp1Cre | PMID:10393122 | MGI:2176467 | Pr. Yumiko Saga (National Institute of Genetics, Japan) |
| Genetic reagent (M. musculus) | Myf5nlacZ | PMID:8918877 | MGI:1857973 | Dr. Shahragim Tajbakhsh (Department of Developmental and Stem Cell Biology, Institut Pasteur, France) |
| Genetic reagent (M. musculus) | Rosa26tdTomato | PMID:20023653 | MGI:3809524 | Dr. Hongkui Zeng (Allen Institute for Brain Science, USA) |
| Genetic reagent (M. musculus) | Rosa26mT/mG | PMID:17868096 | MGI:3716464 | Pr. Philippe Soriano (Icahn School of Medicine at Mt. Sinai, USA) |
| Genetic reagent (M. musculus) | PdgfraH2BGFP | PMID:12748302 | MGI:2663656 | Pr. Philippe Soriano (Icahn School of Medicine at Mt. Sinai, USA) |
| Genetic reagent (M. musculus) | MyodiCre | PMID:19464281 | MGI:3840216 | Pr. David Goldhamer (University of Connecticut, USA) |
| Genetic reagent (M. musculus) | Myf5GFP-P | PMID:15386014 | MGI:3055340 | Dr. Shahragim Tajbakhsh (Department of Developmental and Stem Cell Biology, Institut Pasteur, France) |
| Chemical compound, drug | Sucrose,for molecular biology, ≥ 99.5% (GC) | Sigma-Aldrich | S0389-500G | |
| Chemical compound, drug | Gelatin | Sigma-Aldrich | G-7041 | |
| Antibody | Anti-Foxp2 5C11A8 (Mouse monoclonal) | Santa Cruz | SC-517261 | IF (1:200) |
| Antibody | Anti-β-gal (Chicken polyclonal) | Abcam | Cat. #: ab9361 RRID:AB_307210 | IF (1:1000) |
| Antibody | Anti-β-gal (Rabbit polyclonal) | MP Biomedicals | Cat. #: MP 559761 RRID:AB_2687418 | IF (1:1500) |
| Antibody | Anti-GFP (Chicken polyclonal) | Aves Labs | Cat. #: 1020 RRID:AB_10000240 | IF (1:500) |
| Antibody | Anti-GFP (Chicken polyclonal) | Abcam | Cat. #: 13970 RRID:AB_300798 | IF (1:1000) |
| Antibody | Anti-Myod (Mouse monoclonal) | Dako | Cat. #: M3512 RRID:AB_2148874 | IF (1:100) |
| Antibody | Anti-Myod (Mouse monoclonal) | BD-Biosciences | Cat. #: 554130 RRID:AB_395255 | IF (1:500) |
| Antibody | Anti-Pax7 (Mouse monoclonal) | DSHB | Cat. #: Pax7 RRID:AB_528428 | IF (1:20) |
| Antibody | Anti-Myog (Mouse monoclonal) | DSHB | Cat. #: F5D RRID:AB_2146602 | IF (1:20) |
| Antibody | Alexa Fluor 633 F(ab')2 Fragment of Goat Anti-Rabbit IgG (H + L) (polyclonal antibody) | Life Technologies | Cat. #: A-21072 RRID:AB_2535733 | IF (1:500) |
| Antibody | Alexa Fluor 555 F(ab')2 Fragment of Goat Anti-Rabbit IgG (H + L) (polyclonal antibody) | Life Technologies | Cat. #: A-21430 RRID:AB_2535851 | IF (1:500) |
| Antibody | Alexa Fluor 488 F(ab')2 Fragment of Goat Anti-Rabbit IgG (H + L) (polyclonal antibody) | Life Technologies | Cat. #: A-11070 RRID:AB_2534114 | IF (1:500) |
| Antibody | Alexa Fluor 633 Goat Anti-Chicken IgG (H + L) (polyclonal antibody) | Life Technologies | Cat. #: A-21103 RRID:AB_2535756 | IF (1:500) |
| Antibody | Alexa Fluor 488 Goat Anti-Chicken IgG (H + L) (polyclonal antibody) | Life Technologies | Cat. #: A-11039 RRID:AB_2534096 | IF (1:500) |
| Antibody | Alexa Fluor 633 Goat Anti-Mouse IgG1 (γ1) (polyclonal antibody) | Life Technologies | Cat. #: A-21126 RRID:AB_2535768 | IF (1:500) |
| Antibody | Alexa Fluor488 AffiniPure Goat Anti-Mouse IgG1 (γ1) (polyclonal antibody) | Jackson ImmunoResearch | Cat. #: 115-545-205 RRID:AB_2338854 | IF (1:500) |
| Antibody | Cy3-AffiniPure Goat Anti-Mouse IgG1 (γ1) (polyclonal antibody) | Jackson ImmunoResearch | Cat. #: 115-165-205 RRID:AB_2338694 | IF (1:500) |
| Antibody | Cy3-AffiniPure Goat Anti-Mouse IgG2a (γ2a) (polyclonal antibody) | Jackson ImmunoResearch | Cat. #: 115-165-206 RRID:AB_2338695 | IF (1:500) |
| Antibody | Dylight 405 Goat Anti-Mouse IgG2a (γ2a) (polyclonal antibody) | Jackson ImmunoResearch | Cat. #: 115-475-206 RRID:AB_2338800 | IF (1:500) |
| Commercial assay, kit | Hoechst 33,342 | Thermo Scientific | Cat. #:H3570 | |
| Commercial assay, kit | RNAscope Multiplex Fluorescent reagent Kit-V2 | ACD/Bio-techne | Cat. #: 323100 | |
| Commercial assay, kit | RNAscope H202 & Protease Plus Reagents | ACD/Bio-techne | Cat #: 322330 | |
| Commercial assay, kit | Opal 650 Reagent Pack | PerkinElmer | Cat. #: FP1496001KT | 1:1,500 of reconstituted reagent in RNAscope Multiplex TSA Buffer |
| Commercial assay, kit | Opal 570 Reagent Pack | PerkinElmer | Cat. #: FP1488001KT | 1:1,500 of reconstituted reagent in RNAscope Multiplex TSA Buffer |
| Commercial assay, kit | RNAscope Mm-Pdgfa | Advanced Cell Diagnostics, Inc | Cat #:411361 | |
| Commercial assay, kit | RNAscope Mm-Pdgfra | Advanced Cell Diagnostics, Inc | Cat #:480661-C2 | |
| Commercial assay, kit | RNAscope Mm-Bmpr1b | Advanced Cell Diagnostics, Inc | Cat #:533941 | |
| Commercial assay, kit | RNAscope Mm-Efnb1 | Advanced Cell Diagnostics, Inc | Cat #:526761 | |
| Commercial assay, kit | RNAscope Mm-Bmp4-O1-C3 | Advanced Cell Diagnostics, Inc | Cat #:527501-C3 | |
| Commercial assay, kit | RNAscope Mm-Ephb1-C3 | Advanced Cell Diagnostics, Inc | Cat #:567571-C3 | |
| Commercial assay, kit | RNAscope Mm-Scube1 | Advanced Cell Diagnostics, Inc | Cat #:488131 | |
| Chemical compound, drug | Paraformaldehyde | Electron Microscopy Sciences | Cat. #: 15710 | |
| Chemical compound, drug | Isopentane | VWR | Cat. #: 24872.298 | |
| Chemical compound, drug | Triton X-100 | Sigma | Cat. #: T8787 | |
| Chemical compound, drug | Tween 20 | Sigma | Cat. #: P1379 | |
| Chemical compound, drug | TrypLE | ThermoFisher | Cat #: 12604013 | |
| Chemical compound, drug | Calcein Blue | eBioscience | Cat #: 65-0855-39 | |
| Chemical compound, drug | Propidium Iodide | ThermoFisher | Cat #: P1304MP | |
| Commercial assay, kit | Chromium Next GEM Chip G Single Cell Kit, 16 rxns | 10 X Genomics | Cat #: 1000127 | |
| Commercial assay, kit | Chromium Next GEM Single Cell 3' GEM, Library & Gel Bead Kit v3.1, 4 rxns | 10 X Genomics | Cat #:1000128 | |
| Commercial assay, kit | NextSeq 500/550 High Output Kit v2.5 | Illumina | Cat #: 20024906 | |
| Commercial assay, kit | Agilent High Sensitivity DNA Kit | Agilent | Cat #:5067–4626 | |
| Commercial assay, kit | Agilent High Sensitivity DNA Reagents | Agilent | Cat #:5067–4627 | |
| Commercial assay, kit | Qubit dsDNA HS Assay Kit | Life Technologies | Cat #:Q32854 | |
| Software, algorithm | RStudio | Rstudio | ||
| Software, algorithm | Anaconda | Anaconda Inc | ||
| Software, algorithm | Zen | Zeiss | ||
| Software, algorithm | Cytoscape | Cytoscape Team | ||
| Software, algorithm | Fiji | Johannes Schindelin, Ignacio Arganda-Carreras, Albert Cardona, Mark Longair, Benjamin Schmid, and others | ||
| Software, algorithm | Prism | GraphPad Software | ||
| Software, algorithm | FlowJo | FlowJo |





