Gut microbiota induces high platelet response in patients with ST segment elevation myocardial infarction after ticagrelor treatment
Figures
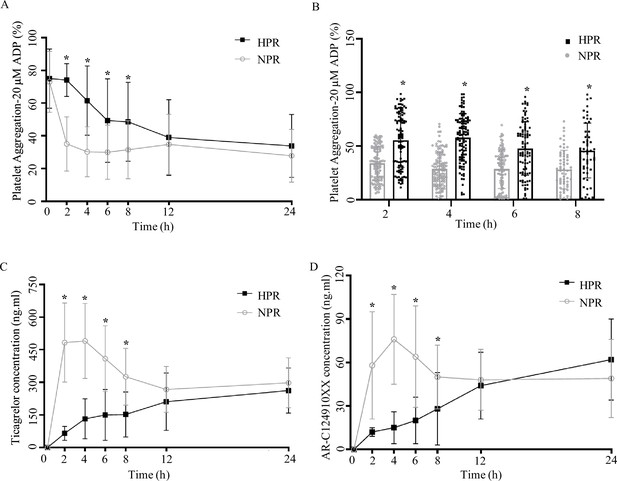
Pharmacodynamic and pharmacokinetic assessment of ticagrelor in the high platelet reactivity (HPR) and normal platelet reactivity (NPR) groups.
(A) Platelet aggregation measured using light transmission aggregometry of line chart at baseline and 2, 4, 6, 8, 12, and 24 hr after the ticagrelor loading dose in patients with NPR and HPR. (B) Platelet aggregation of ticagrelor shown in bar graph at 2, 4, 6, and 8 hr after the ticagrelor loading dose in patients with NPR and HPR. (C, D) The plasma concentration of ticagrelor (C) and its major active metabolite AR-C124910XX (D) during the 24 hr following administration of the loading dose of ticagrelor. Values are expressed as the mean. Error bars indicate standard deviation. *p < 0.05 versus NPR.
-
Figure 1—source data 1
Twelve-month follow-up results of enrolled normal platelet reactivity (NPR) and high platelet reactivity (HPR) patients.
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Datasheet of plasma concentration of ticagrelor and its major active metabolite AR-C124910XX at baseline and 2, 4, 6, 8, 12, and 24 hr following administration of the loading dose of ticagrelor in normal platelet reactivity (NPR) and high platelet reactivity (HPR) patients (related to Figure 1A and B).
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig1-data2-v1.xlsx
-
Figure 1—source data 3
Datasheet of platelet aggregation measured using light transmission aggregometry at baseline and 2, 4, 6, 8, 12, and 24 hr after the ticagrelor loading dose in normal platelet reactivity (NPR) and high platelet reactivity (HPR) patients (related to Figure 1C and D).
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig1-data3-v1.xlsx
-
Figure 1—source data 4
General clinical data of enrolled normal platelet reactivity (NPR) and high platelet reactivity (HPR) patients.
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig1-data4-v1.xlsx
-
Figure 1—source data 5
Comparison of demographic and clinical characteristics between NPR and HPR groups.
NPR, normal platelet reactivity; HPR, high platelet reactivity; MI, myocardial infarction; PCI, percutaneous coronary intervention; CABG, coronary artery bypass grafting; ACE, angiotensin-converting enzyme; PPI, proton pump inhibitor; WBC, white blood cell; FPG, fasting plasma glucose; TC, serum total cholesterol; TG, serum triglyceride; LDL-C, serum low-density lipoprotein cholesterol; HDL-C, serum high-density lipoprotein cholesterol. Data are expressed as the mean ± SD (standard deviation) or n (%), p-value determined by unpaired t-test for continuous variables or Fisher’s exact test for proportions.
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig1-data5-v1.docx
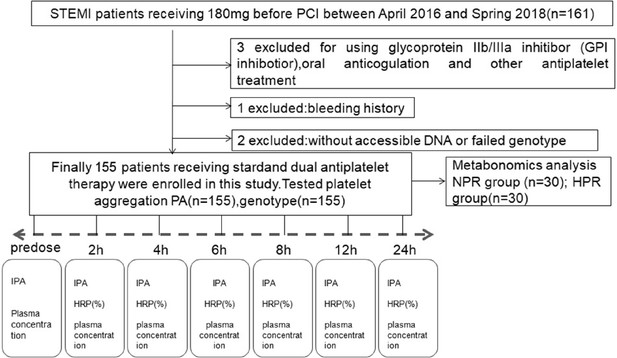
Flowchart illustrating the recruitment of patients based on the exclusion and inclusion criteria.
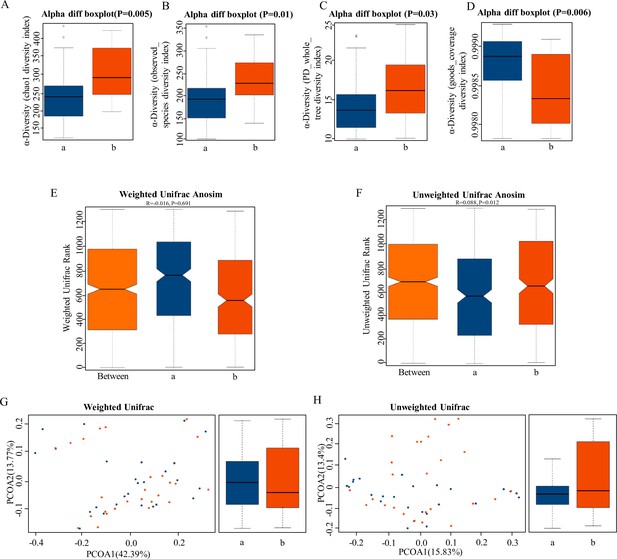
The α-diversity and β-diversity indices of the fecal microbiome in the normal platelet reactivity (NPR) and high platelet reactivity (HPR) groups.
(A, B, C, D) Box plots depict differences in the fecal microbiome diversity indices between the PD and healthy groups according to the chao1 index (A), observed species index (B), PD whole tree index (C), and goods coverage diversity index (D) based on the OTU counts. Each box plot represents the median, interquartile range, minimum, and maximum values. (E, F, G, H) Unweighted and weighted ANOSIMs and PCOA based on the distance matrix of UniFrac dissimilarity of the fecal microbial communities in the HPR and NPR groups. Box and whiskers distribution of the intra-group unweighted UniFrac distances (E) and intra-group weighted UniFrac distances (F) calculated for HPR and NPR groups. Respective ANOSIM R values show the community variation between the compared groups, and significant p-values are indicated, as calculated using Tukey post hoc test after Kruskal-Wallis test for multiple comparisons. The axes represent the two dimensions explaining the greatest proportion of variance in the communities. Each symbol represents a sample, and each line connects a pair of samples. a, NPR group (blue); b, HPR group (red). OTU, operational taxonomic unit; ANOSIM, analyses of similarities; PCOA, principal coordinates analysis.
-
Figure 2—source data 1
Closing report of 16S rDNA amplicon sequencing related to Figure 2.
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig2-data1-v1.pdf
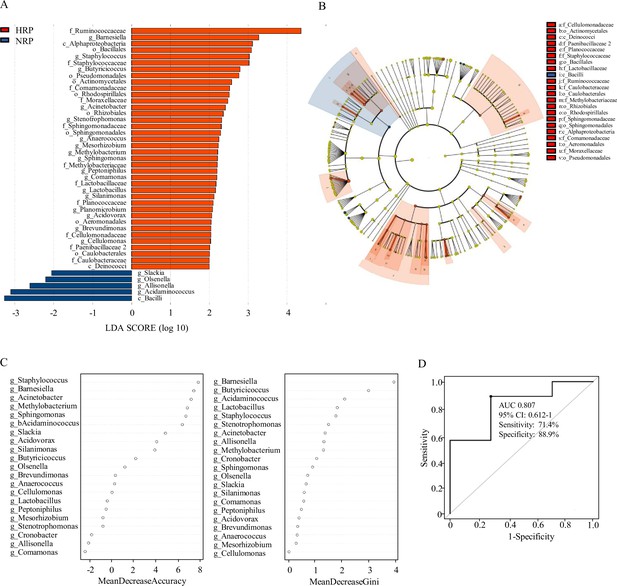
Taxonomic differences of fecal microbiota in the normal platelet reactivity (NPR) and high platelet reactivity (HPR) groups.
(A) Linear discriminant analysis (LDA) effect size (LEfSe) analysis revealed significant bacterial differences in gut microbiota between the NPR (negative score) and HPR (positive score) groups. The LDA scores (log10) >2 and p < 0.05 are listed. (B) Cladogram using LEfSe method indicating the phylogenetic distribution of fecal microbiota associated with HPR and NPR groups. (C) The predictive model based on genus-level abundance taxa using an RF model. The relative importance of each genus in the predictive model was performed using the mean decreasing accuracy and the Gini coefficient for fecal microbiota. (D) ROC curve generated by RF in gut microbiota. The plots shown in the ROC represent the corresponding optimal threshold. RF, random forest; ROC, receiver operating characteristic; AUC, area under the ROC curve; CI, confidence interval.
-
Figure 3—source data 1
Closing report of 16S rDNA amplicon sequencing related to Figure 3.
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig3-data1-v1.pdf
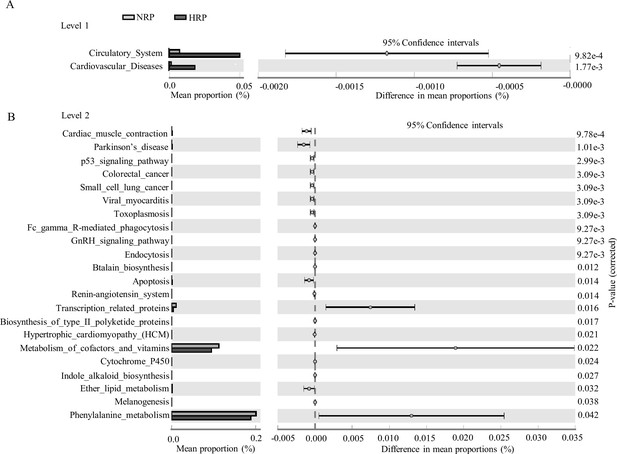
Functional predictions for the fecal microbiome of the normal platelet reactivity (NPR) and high platelet reactivity (HPR) groups.
The important KEGG pathway of gut microbiota in the HPR and NPR groups was identified using stamp software. White’s nonparametric t-test was used to compare the abundance differences between the two groups. The confidence interval was estimated using the percentile bootstrap method (10,000 repetitions). KEGG, Kyoto Encyclopedia of Genes and Genomes; Ko, KEGG homologues; PICRUS, community phylogeny survey by reconstructing unobserved states.
-
Figure 4—source data 1
Relative abundance data for prediction of gut microbiota function at level 2 of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway.
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig4-data1-v1.xls
-
Figure 4—source data 2
Relative abundance data for prediction of gut microbiota function at level 3 of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway.
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig4-data2-v1.xls
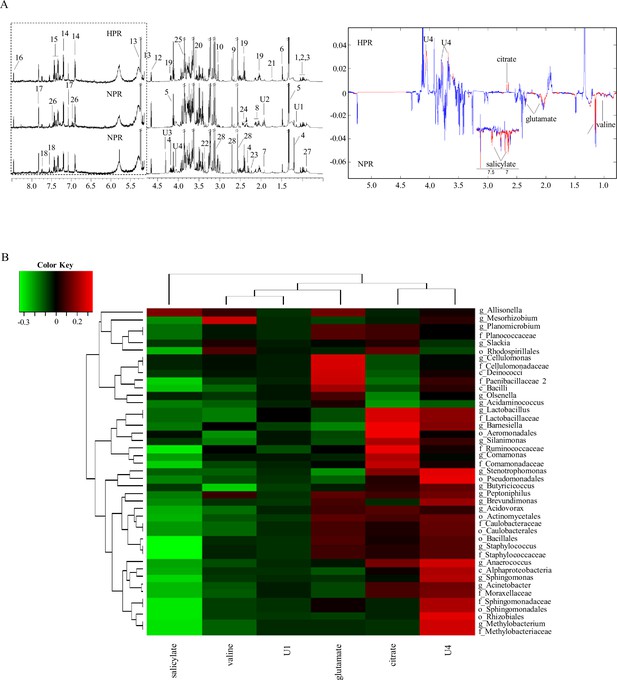
1H NMR spectra of metabolites from the normal platelet reactivity (NPR) and high platelet reactivity (HPR) groups.
(A) NMR spectrum and signal assignment diagram. There were 30 samples in the NPR group and 30 samples in the HPR group. The dotted line on the left is the signal spectrum amplified 30-fold. Each number in the figure represents a metabolite. The right is a two color loading graph of the nonparametric test (univariate analysis). 1, leucine; 2, isoleucine; 3, valine; 4, 3-hydroxybutyric acid; 5, lactate; 6, alanine; 7, acetate; 8, glutamate; 9, citrate; 10, creatine; 11, creatinine; 12, β-glucose; 13, a-glucose; 14, tyrosine; 15, phenylalanine; 16, formate; 17, histidine; 18, tryptophan; 19, pyroglutamate; 20, glycine; 21, lysine; 22, methanol; 23, acetone; 24, succinate; 25, glucitol; 26, salicylate; 27, 2-Hydroxybutyric acid; 28, EDTA; U1, unknown 1; U2, unknown 2; U3, unknown 3; U4, unknown 4. (B) Correlation analysis between 16S and significantly changed metabolites. The intensity of the color represents the r value (correlation) (negative score, green; positive score, red).
-
Figure 5—source code 1
The original code file of 1H NMR serum metabolite analysis in normal platelet reactivity (NPR) and high platelet reactivity (HPR) patients.
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig5-code1-v1.zip
-
Figure 5—source data 1
Linear discriminant analysis effect size (LEfSe) analysis was used to analyze the correlation between gut microbiota and metabolites in normal platelet reactivity (NPR) and high platelet reactivity (HPR) patients.
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig5-data1-v1.xlsx
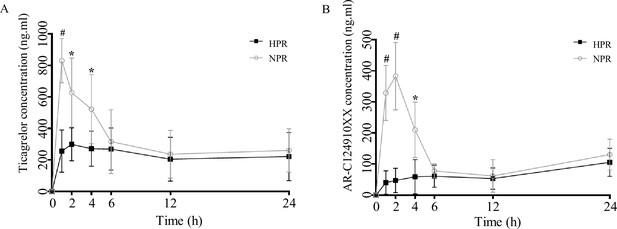
Post-transplanted pharmacokinetic assessment of ticagrelor in recipient mice of normal platelet reactivity (NPR) and high platelet reactivity (HPR) groups.
The plasma concentration of ticagrelor (A) and its major active metabolite AR-C124910XX (B) during the 24 hr following administration of ticagrelor loading dose in fecal microbiota transplantation mice. n = 6; values are expressed as the mean. Error bars indicate standard deviation. *p < 0.05 versus NPR, #p < 0.01 versus NPR.
-
Figure 6—source data 1
Datasheet of plasma concentration of ticagrelor and its major active metabolite AR-C124910XX in fecal microbiota transplantation mice at baseline and 1, 2, 4, 6, 12, and 24 hr after the ticagrelor loading dose.
- https://cdn.elifesciences.org/articles/70240/elife-70240-fig6-data1-v1.xlsx
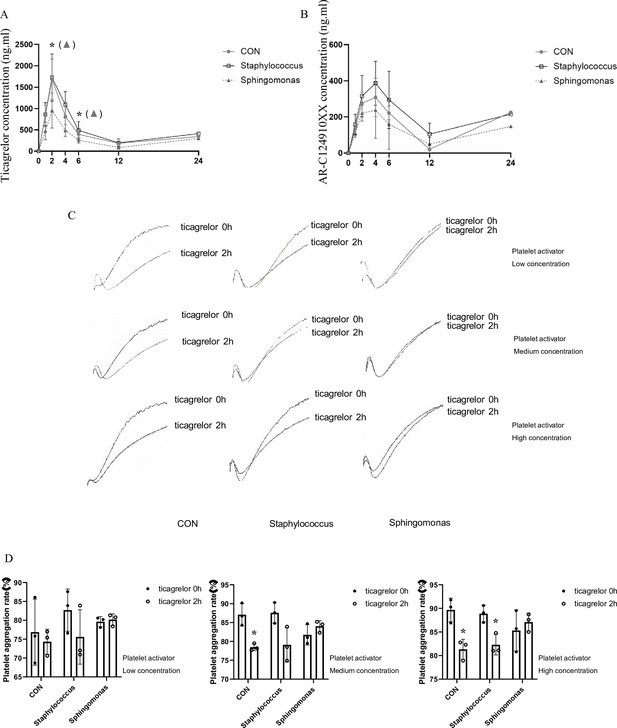
Effects of intragastric administration of Staphylococcus and Sphingomonas on pharmacokinetic assessment of ticagrelor and platelet aggregation.
Suspensions of living Staphylococcus and Sphingomonas were administrated by oral gavage to C57BL/6 mice three times per week at a dose of 5 × 108 CFUs/0.1 mL PBS for 5 weeks. Sterile PBS was used as a control. (A, B) Plasma concentration of ticagrelor (A) and its major active metabolite AR-C124910XX (B) during the 24 hr following administration of the loading dose of ticagrelor. Values are expressed as mean. Error bars indicate standard deviation. *p < 0.05 versus CON. (C) Aggregation traces of washed platelets treated with activator were measured by aggregometry. (D) Maximum platelet aggregation rate with different concentrations of activator (n = 3, values are expressed as means. Error bars indicate standard deviation. *p < 0.05 versus 0 hr).
-
Appendix 1—figure 1—source data 1
Datasheet of plasma concentration of ticagrelor and its major active metabolite AR-C124910XX in single bacterial genera gavaged mice at baseline and 1, 2, 4, 6, 12, and 24 hr after the ticagrelor loading dose (related to Appendix 1—figure 1A, B).
- https://cdn.elifesciences.org/articles/70240/elife-70240-app1-fig1-data1-v1.xlsx
-
Appendix 1—figure 1—source data 2
Screenshot of original data of platelet aggregation measured using light transmission aggregometry in single bacterial genera gavaged mice at baseline and 1, 2, 4, 6, 12, and 24 hr after the ticagrelor loading dose (Part 1).
(related to Appendix 1—figure 1C).
- https://cdn.elifesciences.org/articles/70240/elife-70240-app1-fig1-data2-v1.doc
-
Appendix 1—figure 1—source data 3
Screenshot of original data of platelet aggregation measured using light transmission aggregometry in single bacterial genera gavaged mice at baseline and 1, 2, 4, 6, 12, and 24 hr after the ticagrelor loading dose (Part 2) (related to Appendix 1—figure 1C).
- https://cdn.elifesciences.org/articles/70240/elife-70240-app1-fig1-data3-v1.doc
-
Appendix 1—figure 1—source data 4
Datasheet of platelet aggregation measured using light transmission aggregometry in single bacterial genera gavaged mice at baseline and 1, 2, 4, 6, 12, and 24 hr after the ticagrelor loading dose (related to Appendix 1—figure 1D).
- https://cdn.elifesciences.org/articles/70240/elife-70240-app1-fig1-data4-v1.xlsx
Tables
Genotype and allele distributions for the three polymorphisms of ABCB1 in NPR and HPR groups.
| Genotype/ allele | NPR (%) | HWE-P | HPR (%) | HWE-P | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| Rs1045642 | ||||||
| CC | 24 (26.7) | 0.099 | 24 (36.9) | 0.723 | 1 | |
| CT | 53 (58.9) | 30 (46.2) | 0.566 (0.285–1.192) | 0.121 | ||
| TT | 13 (14.4) | 11 (16.9) | 0.846 (0.332–2.320) | 0.739 | ||
| C allele | 101 (56.1) | 78 (60.0) | 1 | |||
| T allele | 79 (43.9) | 52 (40.0) | 0.852 (0.532–1.351) | 0.494 | ||
| Rs2032582 | ||||||
| GG | 15 (16.7) | 0.360 | 9 (13.8) | 0.111 | 1 | |
| GT | 34 (37.8) | 29 (44.6) | 1.422 (0.560–3.567) | 0.473 | ||
| GA | 11 (12.2) | 9 (13.8) | 1.364 (0.435–4.374) | 0.614 | ||
| TT | 14 (15.6) | 7 (10.7) | 0.833 (0.238–2.678) | 0.771 | ||
| TA | 12 (13.3) | 8 (12.3) | 1.111 (0.339–3.578) | 0.865 | ||
| AA | 4 (4.4) | 3 (4.6) | 1.250 (0.265–5.728) | 0.798 | ||
| G allele | 75 (41.7) | 56 (43.1) | 1 | |||
| T allele | 74 (41.1) | 51 (39.2) | 0.923 (0.568–1.495) | 0.752 | ||
| A allele | 31 (17.2) | 23 (17.7) | 0.994 (0.511–1.874) | 0.985 | ||
| Rs1128503 | ||||||
| CC | 6 (6.7) | 0.670 | 8 (12.3) | 0.662 | 1 | |
| CT | 51 (56.7) | 28 (43.1) | 0.412 (0.123–1.243) | 0.125 | ||
| TT | 33 (36.7) | 29 (44.6) | 0.659 (0.191–1.955) | 0.483 | ||
| C allele | 63 (35.0) | 44 (33.8) | 1 | |||
| T allele | 117 (65.0) | 86 (66.2) | 1.052 (0.661–1.700) | 0.833 | ||
-
Table 1—source code 1
The original code file of sequencing analysis for tagSNPs of ATP binding cassette subfamily B member 1 (ABCB1) gene in normal platelet reactivity (NPR) and high platelet reactivity (HPR) patients (related to Table 1).
- https://cdn.elifesciences.org/articles/70240/elife-70240-table1-code1-v1.zip
-
Table 1—source data 1
ATP binding cassette subfamily B member 1 (ABCB1) tagSNPs from the HapMap database and primer sequences used in genotyping analysis for ABCB1.
MAF, miner allele frequence.
- https://cdn.elifesciences.org/articles/70240/elife-70240-table1-data1-v1.docx
Significant correlations between the differential fecal metabolites and microbes in the class, order, family, and genus levels.
Statistical method: Pearson’s correlation coefficient; listed correlation coefficients are those with p-value < 0.05.
| Glutamate | Citrate | Salicylate | |
|---|---|---|---|
| Class | |||
| c_Deinococci | 0.2759636 | – | – |
| Order | |||
| o_Aeromonadales | – | 0.3212488 | – |
| o_Bacillales | – | – | –0.343397 |
| o_Rhizobiales | – | – | –0.307195 |
| o_Sphingomonadales | – | – | –0.315105 |
| Family | |||
| f_Cellulomonadaceae | 0.2813448 | – | – |
| f_Lactobacillaceae | –0.099515 | 0.2799663 | – |
| f_Methylobacteriaceae | – | – | –0.305264 |
| f_Ruminococcaceae | – | 0.3119055 | –0.314255 |
| f_Sphingomonadaceae | – | – | –0.315105 |
| f_Staphylococcaceae | – | – | –0.336272 |
| Genus | |||
| g_Barnesiella | – | 0.3164411 | – |
| g_Cellulomonas | 0.2813448 | – | – |
| g_Lactobacillus | – | 0.2799663 | – |
| g_Methylobacterium | – | – | –0.305264 |
| g_Sphingomonas | – | – | –0.281848 |
| g_Staphylococcus | – | – | –0.336272 |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/70240/elife-70240-transrepform1-v1.docx
-
Source code 1
The original code file of 16S rRNA gene sequencing of gut microbiota c (Part 1).
- https://cdn.elifesciences.org/articles/70240/elife-70240-code1-v1.zip
-
Source code 2
The original code file of 16S rRNA gene sequencing of gut microbiota in normal platelet reactivity (NPR) and high platelet reactivity (HPR) patients (Part 2).
- https://cdn.elifesciences.org/articles/70240/elife-70240-code2-v1.zip
-
Source code 3
The original code file of 16S rRNA gene sequencing of gut microbiota in normal platelet reactivity (NPR) and high platelet reactivity (HPR) patients (Part 3).
- https://cdn.elifesciences.org/articles/70240/elife-70240-code3-v1.zip
-
Source code 4
The original code file of 16S rRNA gene sequencing of gut microbiota in normal platelet reactivity (NPR) and high platelet reactivity (HPR) patients (Part 4).
- https://cdn.elifesciences.org/articles/70240/elife-70240-code4-v1.zip
-
Reporting standard 1
STROBE statement.
- https://cdn.elifesciences.org/articles/70240/elife-70240-repstand1-v1.doc
-
Appendix 1—figure 1—source data 1
Datasheet of plasma concentration of ticagrelor and its major active metabolite AR-C124910XX in single bacterial genera gavaged mice at baseline and 1, 2, 4, 6, 12, and 24 hr after the ticagrelor loading dose (related to Appendix 1—figure 1A, B).
- https://cdn.elifesciences.org/articles/70240/elife-70240-app1-fig1-data1-v1.xlsx
-
Appendix 1—figure 1—source data 2
Screenshot of original data of platelet aggregation measured using light transmission aggregometry in single bacterial genera gavaged mice at baseline and 1, 2, 4, 6, 12, and 24 hr after the ticagrelor loading dose (Part 1).
(related to Appendix 1—figure 1C).
- https://cdn.elifesciences.org/articles/70240/elife-70240-app1-fig1-data2-v1.doc
-
Appendix 1—figure 1—source data 3
Screenshot of original data of platelet aggregation measured using light transmission aggregometry in single bacterial genera gavaged mice at baseline and 1, 2, 4, 6, 12, and 24 hr after the ticagrelor loading dose (Part 2) (related to Appendix 1—figure 1C).
- https://cdn.elifesciences.org/articles/70240/elife-70240-app1-fig1-data3-v1.doc
-
Appendix 1—figure 1—source data 4
Datasheet of platelet aggregation measured using light transmission aggregometry in single bacterial genera gavaged mice at baseline and 1, 2, 4, 6, 12, and 24 hr after the ticagrelor loading dose (related to Appendix 1—figure 1D).
- https://cdn.elifesciences.org/articles/70240/elife-70240-app1-fig1-data4-v1.xlsx





