Ca2+/CaM binding to CaMKI promotes IMA-3 importin binding and nuclear translocation in sensory neurons to control behavioral adaptation
Figures
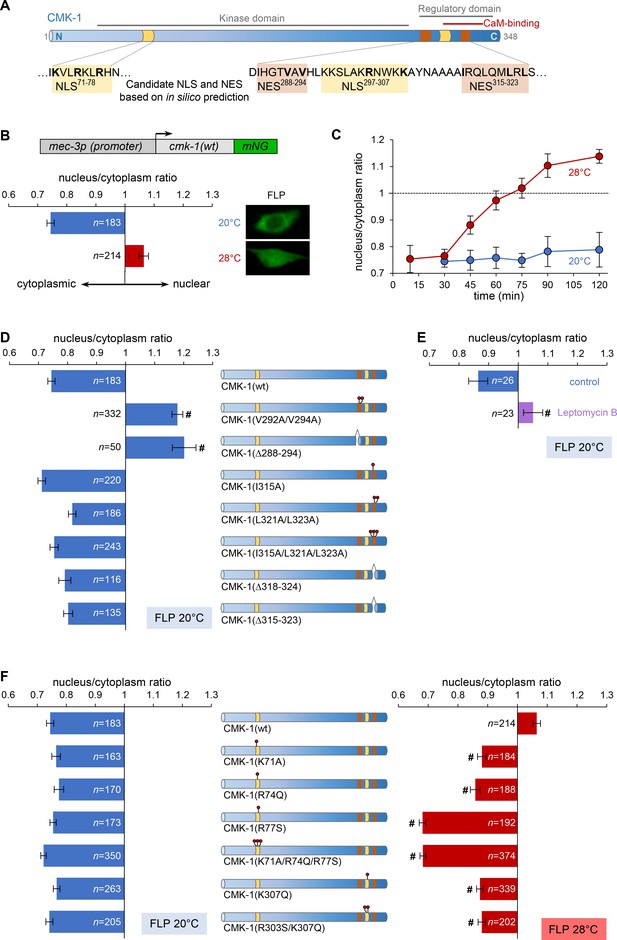
Specific nuclear export sequence (NES) and nuclear localization signal (NLS) control CMK-1 localization in response to temperature in FLP.
(A) Schematic of CMK-1 topology highlighting the localization of in silico-predicted NES and NLS and their sequence. (B) Subcellular localization of CMK-1(wt)::mNeonGreen (mNG) reporter expressed in FLP via the depicted transgene (top). Average nuclear/cytoplasm fluorescent signal ratio (± SEM, left) and representative confocal micrographs (right) showing heat-evoked nuclear translocation of wild-type CMK-1 in young adult FLP neurons after 90 min at 28°C as compared to control at 20°C. (C) Kinetics of CMK-1::mNG nuclear accumulation. Data as nuclear/cytoplasmic fluorescent signal ratio average (± SEM). (D, E) Subcellular localization of CMK-1::mNG reporters carrying the depicted mutations in candidate NES (D), as well as following 90 min incubation with 50 µM leptomycin B or vehicle control (E). Data as nuclear/cytoplasmic signal ratio average (± SEM). (F) Same as for panel (D), but with mutations in the depicted candidate NLS in animals incubated 90 min at 20°C (blue, left panel) or 90 min at 28°C (red, right panel). #p<0.001 versus CMK-1(wt) by Bonferroni contrasts. The number of animals scored in each condition is indicated in the figure (n). Experiments reported in panels (B), (D), and (F) were run in parallel and the CMK-1(wt) dataset is common across these panels.
-
Figure 1—source data 1
Summary statistics and raw data for Figure 1.
- https://cdn.elifesciences.org/articles/71443/elife-71443-fig1-data1-v2.xlsx
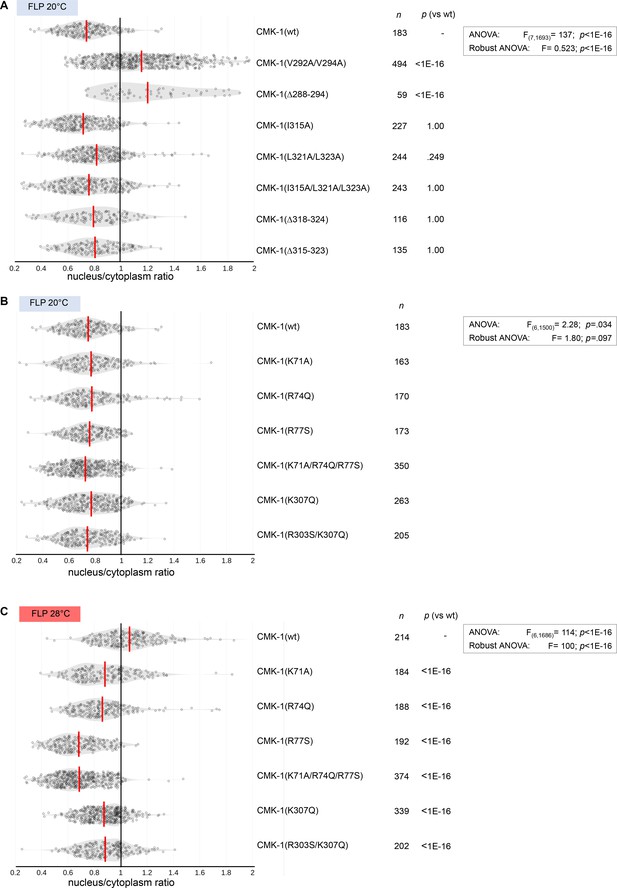
CMK-1 localization data distribution in mutants for nuclear export sequence (NES) and nuclear localization signal (NLS) candidates.
Violin plots with superimposed datapoints depicting the distribution of nucleus/cytoplasmic ratios of the indicated CMK-1::mNG reporters. Red bars: average. n: number of observations. p-Values obtained with Bonferroni post-hoc tests. (A) Data corresponding to Figure 1D. (B) Data corresponding to Figure 1F, left. (C) Data corresponding to Figure 1F, right.
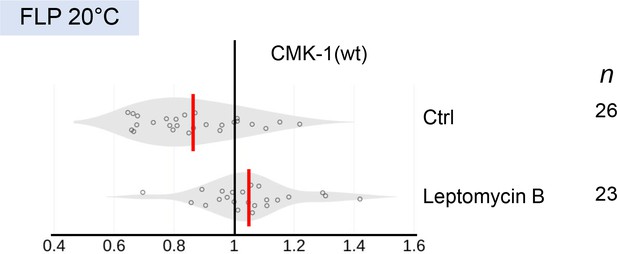
CMK-1 localization data distribution upon leptomycin B treatment.
Violin plots with superimposed datapoints depicting the distributions of nucleus/cytoplasmic ratios of CMK-1(wt)::mNG reporter. Red bars: average. n: number of observations. Data corresponding to Figure 1E.
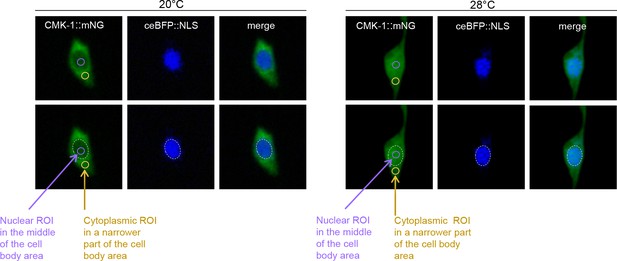
Illustration of regions of interest (ROI) definition.
FLP cell body confocal micrography examples from transgenic animals coexpressing CMK-1::mNG reporter (green channel) and the ceBFP::NLS nuclear marker (blue channel) and incubated at the indicated temperature for 90 min. Dotted ellipse: nucleus shapes. Purple ellipse: nuclear ROI. Yellow ellipse: cytoplasmic ROI.
Intact cell-autonomous calcium signaling and CMK-1 Ca2+/CaM binding ability are essential to regulate CMK-1 subcellular localization in FLP.
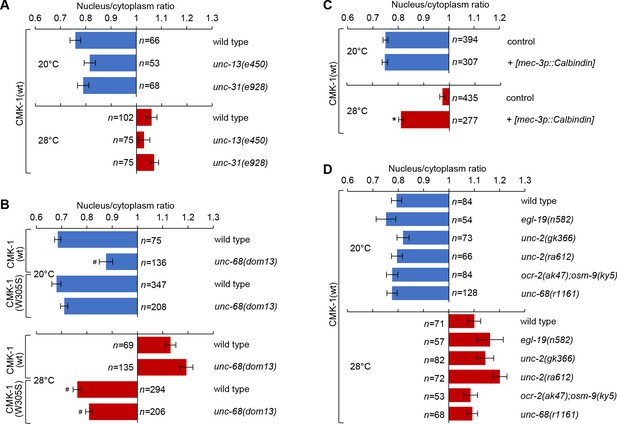
Subcellular localization of CMK-1(wt)::mNG reporters in FLP neurons of young adults of the indicated genotype, maintained 90 min at 20 or 28°C. Data as nuclear/cytoplasmic signal ratio average (± SEM). (A) Comparison showing no significant difference between wild-type and mutants affecting synaptic neurotransmission (unc-13(e540)) or dense core vesicle release (unc-31(e928)). (B) Subcellular localization of CMK-1(wt)::mNG and CMK-1(W305S)::mNG in wild-type and unc-68(dom13) mutant background. The unc-68 gain-of-function mutation reduces the cytoplasmic accumulation of CMK-1(wt) at 20°C, but fails to do so when the CaM binding site is altered in CMK-1(W305S). n ≥ 135 animals. *p<0.001 versus wild-type; #p<0.001 versus CMK-1(wt) for the corresponding condition by Bonferroni contrasts. (C) Subcellular localization of CMK-1(wt)::mNG reporter in [mec-3p::Calbindin] transgenic animals, as well as non-transgenic animals coming from the same growth plates (control). The expression of the Calbindin calcium buffer inhibits CMK-1 nuclear translocation. n ≥ 277. *p<0.001 versus control at the same temperature by Bonferroni contrasts. (D) Same analysis as in (A), with indicated mutants affecting different calcium channels, but showing no statistically significant difference by Bonferroni contrasts.
-
Figure 2—source data 1
Summary statistics and raw data for Figure 2.
- https://cdn.elifesciences.org/articles/71443/elife-71443-fig2-data1-v2.xlsx
CMK-1 localization data distributions and statistical test results.
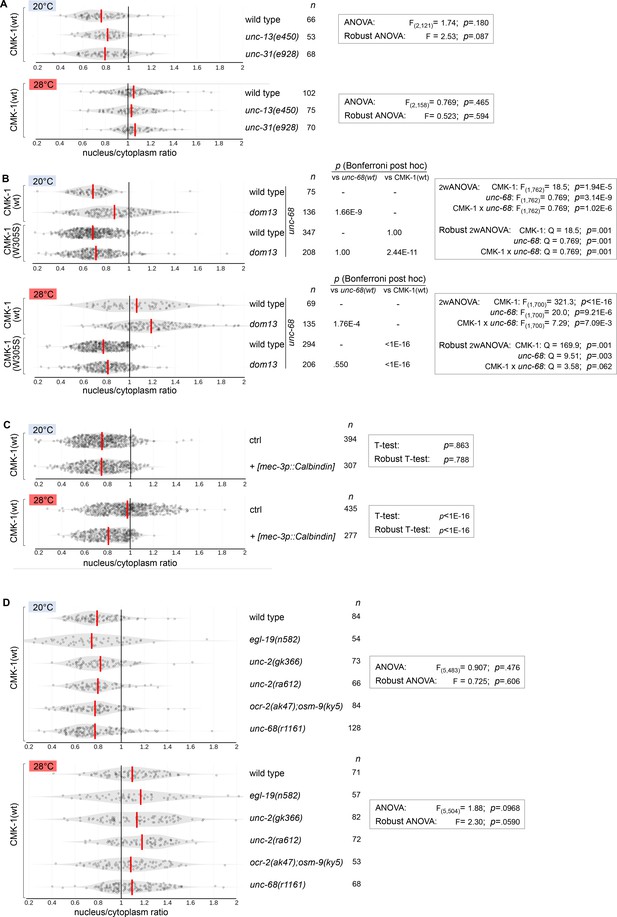
Violin plots with superimposed datapoints depicting the distributions of nucleus/cytoplasmic ratios of the indicated CMK-1::mNG reporters. Red bars: average. n: number of observations. p-Values obtained with Bonferroni post-hoc tests. (A) Data corresponding to Figure 2A. (B) Data corresponding to Figure 2B. (C) Data corresponding to Figure 2C. (D) Data corresponding to Figure 2D.
FLP calcium concentration in Calbindin-expressing transgenic animals and calcium channel mutants.
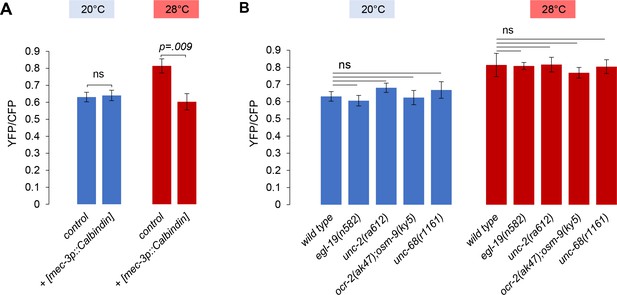
Calcium levels in FLP cell bodies of [egl-46p:.YC2.3] animals incubated at 20 or 28°C for 60 min. Average YFP/CFP ratio (± SEM) of n ≥ 10 animals per conditions. (A) Comparison between wild-type and Calbindin-expressing transgenic animals showing a significant decrease in temperature-evoked calcium elevation. p-Value by Student’s t-test. (B) Comparison between wild-type and different calcium channel mutants showing no significant effect of the mutations. ns, not significant.
Parvalbumin expression in FLP reduces CMK-1 nuclear accumulation.
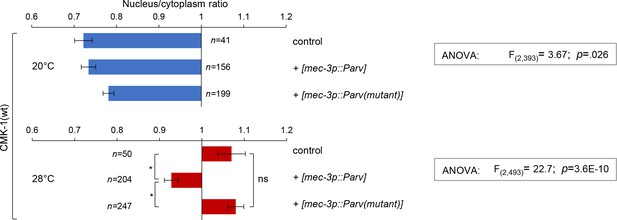
Subcellular localization of CMK-1(wt)::mNG reporters in FLP neurons of young adults of the indicated genotype, incubated 90 min at 20 or 28°C. Data as nuclear/cytoplasmic signal ratio average (± SEM). The number of animals scored in each condition is indicated in the figure (n). *p<0.01 by Bonferroni post-hoc test. ns, not significant.
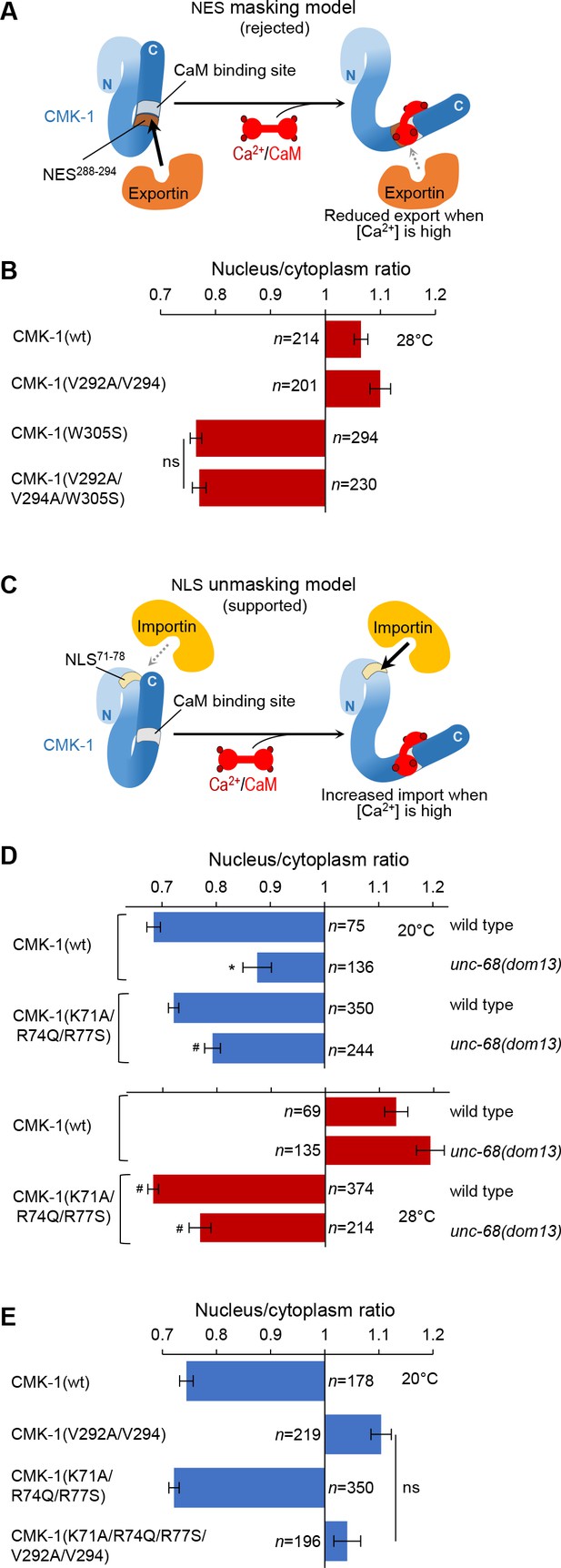
In vivo tests for nuclear export sequence (NES) masking and nuclear localization signal (NLS) unmasking models.
(A) Schematic of the rejected model in which NES288-294 would be masked by Ca2+/CaM binding. (B) Subcellular localization of CMK-1::mNG reporters carrying the indicated mutations in the FLP neurons of animals exposed for 90 min at 28°C. Data as nuclear/cytoplasmic signal ratio average (± SEM), showing that the CaM-binding-disrupting mutation W305S prevents nuclear localization independently of the NES288-294 element. (C) Schematic of the retained model in which NLS71-78 would be unmasked by Ca2+/CaM binding. (D) Subcellular localization of CMK-1(wt)::mNG and the NLS71-78-disrupting mutant CMK-1(K71A/R74Q/R77S)::mNG at 20 and 28°C in both wild-type and unc-68(dom13) backgrounds. Data as nuclear/cytoplasmic signal ratio average (± SEM), showing that the NLS71-78-disrupting mutations counteract the impact of the unc-68 gain-of-function mutation. *p<0.001 versus wild-type; #p<0.001 versus CMK-1(wt) for the corresponding condition by Bonferroni post-hoc tests. Experiments were run in parallel to those reported in Figure 2B, and CMK-1(wt) data are common across the two figures. (E) Subcellular localization of CMK-1::mNG reporters carrying the indicated mutations disrupting either NES288-294, NLS71-78, or both of them. Data as nuclear/cytoplasmic signal ratio average (± SEM), showing no effect of NLS71-78 disruption at 20°C, even when the NES288-294 is impaired. ns, not significant.
-
Figure 3—source data 1
Summary statistics and raw data for Figure 3.
- https://cdn.elifesciences.org/articles/71443/elife-71443-fig3-data1-v2.xlsx
CMK-1 localization data distributions and statistical test results.
Violin plots with superimposed datapoints depicting the distributions of nucleus/cytoplasmic ratios of the indicated CMK-1::mNG reporters. Red bars: average. n: number of observations. p-Values obtained with Bonferroni post-hoc tests. (A) Data corresponding to Figure 3B. (B) Data corresponding to Figure 3D. (C) Data corresponding to Figure 3E.
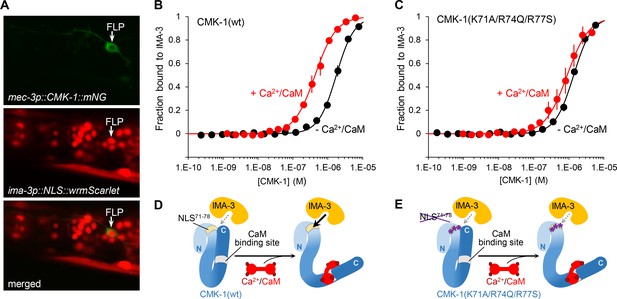
Ca2+/CaM binding to CMK-1 enhances the affinity for IMA-3 importin-α in a NLS71-78-dependent manner.
(A) ima-3 expression analysis with a transcriptional reporter. Representative confocal micrographs showing fluorescence signals in the head of an adult [mec-3p::CMK-1::mNG; ima-3p::NLS::wrmScarlet] transgenic animal. FLP cytoplasm is labeled in green (top). The nuclei of ima-3-expressing cells are labeled in red (middle). FLP is among the cells expressing ima-3p (bottom). (B) Binding curves for the CMK-1(wt)/IMA-3 interaction in the presence (red curve) or the absence (black curve) of Ca2+/CaM, as determined by MicroScale Thermophoresis (see Materials and methods). Data as average of three replicates (± SEM). (C) Same analysis as in (B), but with CMK-1 (K71A/R74Q/R77S) mutant. Kd derived from fitting curves are reported in Table 1. (D, E) Schematic protein interaction models illustrating the increased affinity for IMA-3 when Ca2+/CaM binds CMK-1(wt) (D) and the impact of NLS71-78 mutations, which prevent CaM-dependent IMA-3 affinity increase.
-
Figure 4—source data 1
Summary statistics and raw data for Figure 4.
- https://cdn.elifesciences.org/articles/71443/elife-71443-fig4-data1-v2.xlsx
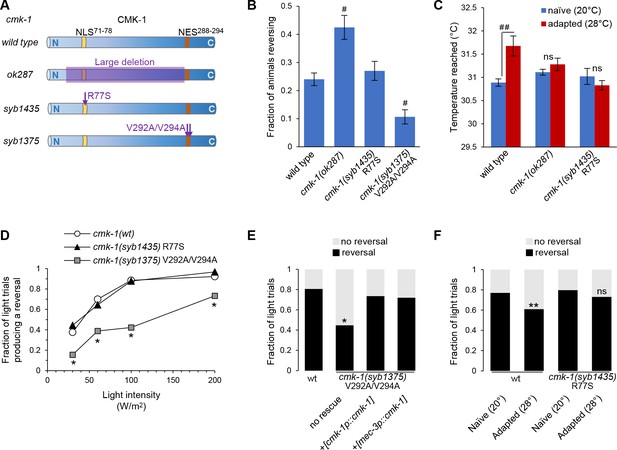
Engineered mutations affecting CMK-1 NLS71-78 and NES288-294 impact the adaptation of FLP-dependent behavior.
(A) Schematic of CMK-1 protein highlighting the position of the deletion in ok287 mutant and of the single-point mutations in syb1435 and syb1375 CRISPR/Cas9 engineered alleles affecting NLS71-78 and NES288-294, respectively. (B) Avoidance response to 4 s heat pulses (at 0.3–0.6 W /m2, see Materials and methods) in adult animals of the indicated genotypes (average ± SEM, n ≥ 21 plates, each scoring at least 50 worms). #p<0.001 versus wild-type by Bonferroni contrasts. (C) Noxious heat thermogradient assays with animals of the indicated genotype maintained at 20°C (blue) or adapted for 1 hr at 28°C (red). Dispersal of animals reported as the temperature corresponding to the third quartile of the worm distribution in the spatial thermogradient. Vertical axis minimum was set to the lowest temperature in the thermogradient (29.5°C). Data as average (± SEM) of n ≥ 12 assays, each scoring more than 100 animals. ##p<0.001 by Bonferroni contrasts. ns, not significant. (D) FLP optogenetic analysis: light dose–response curves in young adult [FLP::CoChR] animals in response to 0.5 s light stimuli. Data as fraction of trials producing a response (n = 90 trials per genotype) showing a reduced response in cmk-1(syb1375) with constitutive nuclear CMK-1 expression. *p<0.001 versus cmk-1(wt) by Fisher’s exact tests. (E) FLP optogenetic analysis: light avoidance rescue experiment in cmk-1(syb1375) mutant. Data acquired as in (D), using 100 W/m2 light stimuli, and expressed as fraction of trials producing reversal or not. n ≥ 100 trials per genotype, each aggregating data from three independent rescue lines. *p<0.01 versus wild-type by Fisher’s exact tests. Transgenes containing cmk-1p and mec-3p promoter both produced a rescue effect. (F) FLP optogenetic analysis: impact of heat adaptation. Data as in (E), in animals maintained at 20°C (naïve) or incubated for 90 min at 28°C (adapted), showing impaired adaptation in cmk-1(syb1475) mutants. n ≥ 120 light trials per condition. **p<0.01 versus naïve control by Fisher’s exact test. ns, not significant.
-
Figure 5—source data 1
Summary statistics and raw data for Figure 5.
- https://cdn.elifesciences.org/articles/71443/elife-71443-fig5-data1-v2.xlsx
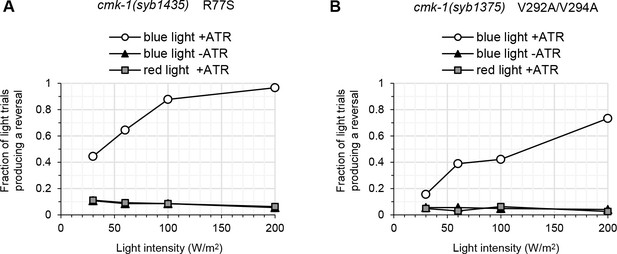
Optogenetic experiment controls.
(A, B) FLP optogenetic analysis: light dose–response curves in young adult [FLP::CoChR] animals in response to 0.5 s light stimuli. Data as fraction of trials producing a response (n = 120 trials per condition). No reversal response above the spontaneous reversal rate is seen for red light stimuli or for blue light stimuli in the absence of ATR.
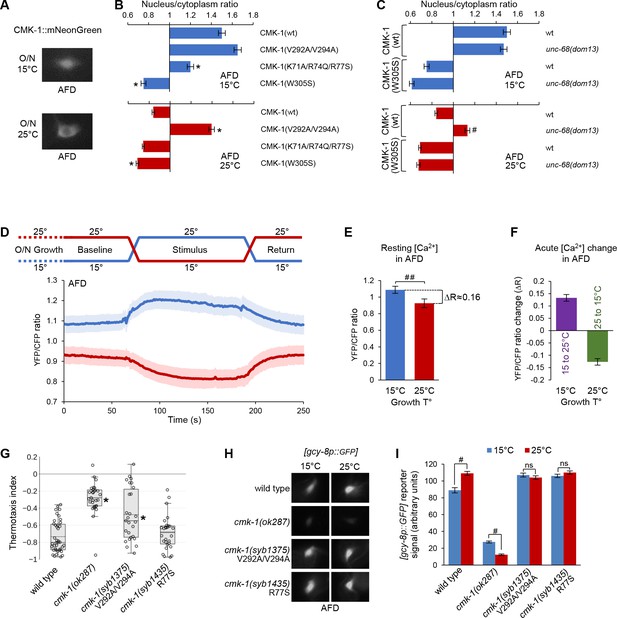
CMK-1 subcellular localization control and function in AFD.
(A) Representative epifluorescence micrographs showing the subcellular expression pattern of CMK-1(wt)::mNG in AFD neurons of young adult animals maintained overnight at 15°C or at 25°C. (B, C) Subcellular localization of wild-type and indicated CMK-1::mNG mutants at 15°C (blue) and at 25°C (red) in wild-type and unc-68(dom13) background. Data as nuclear/cytoplasmic signal ratio average (± SEM) of n ≥ 125 animals. *p<0.01 versus CMK-1(wt); #p<0.01 versus same genotype at 20°C by Bonferroni contrasts. (D) Calcium levels in AFD cell bodies of [ttx-1p:.YC2.3] animals grown overnight (O/N) at 15 or 25°C and submitted to a 2 min temperature increase to 25°C or decrease to 15°C, respectively. Average calcium traces (blue and red lines) with SEM as lighter shades. n ≥ 16 animals. Note the impact of growth temperature on resting calcium levels (baseline and return periods). (E) Resting calcium-level quantification over the baseline period of data in panel (D), highlighting the impact of growth temperature (T°) (average ± SEM, n ≥ 16 animals). ##p<0.01 by Student’s t-test. (F) Acute calcium-level changes caused by the indicated thermal up-steps or down-steps, quantified from data in panel (D) (average ± SEM, n ≥ 16 animals). (G) Thermotaxis assay result in young adult animals of the indicated genotypes grown at 20°C. n ≥ 25 assays, each scoring at least 80 animals. *p<0.01 versus wild-type by Bonferroni post-hoc tests. (H) Representative epifluorescence micrographs of young adult animals expressing a [gcy-8p::GPF] reporter in AFD neurons. Animals of the indicated genotypes were grown either at 15 or 25°C. n ≥ 80 animals per condition. (I) Quantification of GFP reporter signal from the data described in panel (H) (averages ± SEM). ##p<0.001 by Bonferroni contrasts. ns, not significant.
-
Figure 6—source data 1
Summary statistics and raw data for Figure 6.
- https://cdn.elifesciences.org/articles/71443/elife-71443-fig6-data1-v2.xlsx
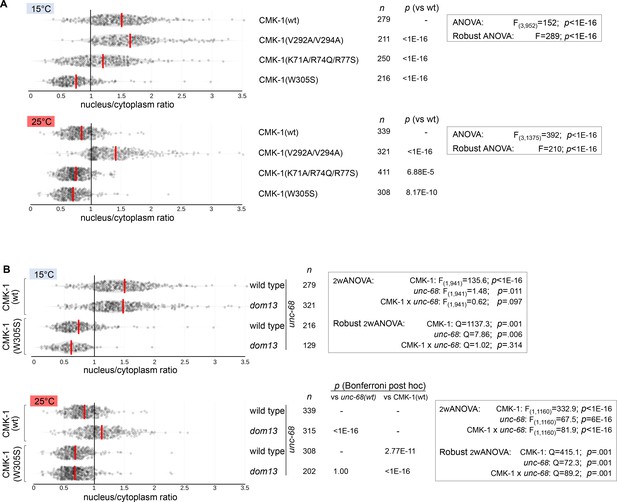
CMK-1 localization data distributions in AFD and statistical test results.
Violin plots with superimposed datapoints depicting the distributions of nucleus/cytoplasmic ratios of the indicated CMK-1::mNG reporters. Red bars: average. n: number of observations. p-Values obtained with Bonferroni post-hoc tests. (A) Data corresponding to Figure 6B. (B) Data corresponding to Figure 6C.
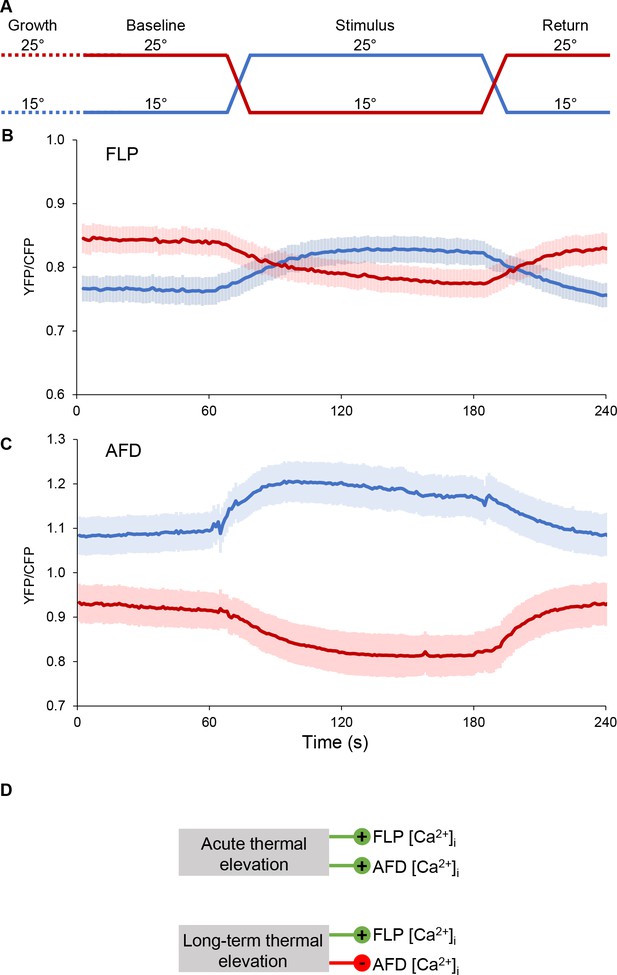
Side-by-side comparison of FLP and AFD calcium response to temperature in the 15–25°C range.
(A) Schematic of the temperature stimulation protocol that was applied to young adult animals grown at 15°C (blue line) or at 25°C (red line). (B, C) Calcium levels recorded in FLP cell bodies of [egl-46p:.YC2.3] animals (n ≥ 6 animals, B) or in AFD cell bodies of [ttx-1p:.YC2.3] animals (n ≥ 16 animals, C). Average calcium traces (blue and red lines) with SEM as lighter shades. AFD data are the same as the one reported in Figure 6D. (D) Schematic summary of the temperature impact on the intracellular calcium concentration ([Ca2+]i) in FLP and AFD. Whereas acute thermal elevations (over a time window of a few minutes) have the same impact in both FLP and AFD, by causing calcium increases, prolonged thermal elevations (over a time window of several hours) have an opposite impact in both neurons, causing long-term calcium increase in FLP, but long-term calcium decrease in AFD.
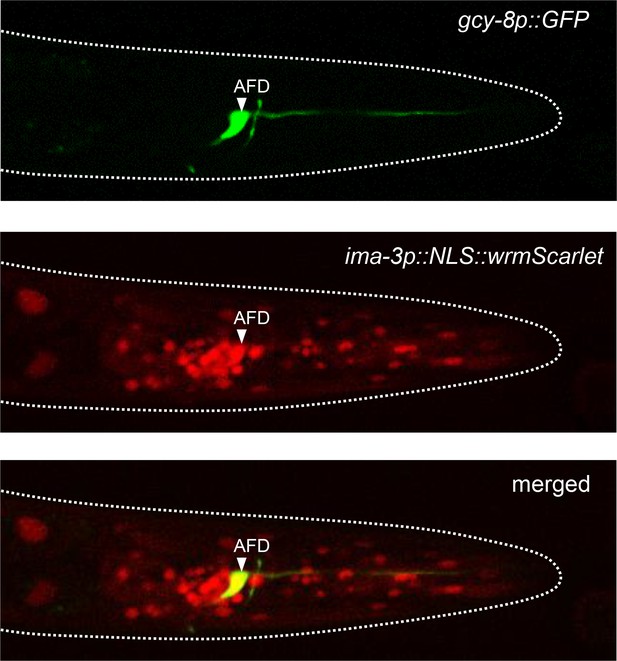
Expression of ima-3 in AFD.
Representative confocal micrographs showing fluorescence signals in the head of an adult [gcy-8::GFP; ima-3p::NLS::wrmScarlet] transgenic animal. AFD is labeled in green (top). The nuclei of ima-3-expressing cells are labeled in red (middle). AFD is among the cells expressing ima-3p (bottom). Dotted lines indicate the animal contour.
Tables
In vitro interaction between CMK-1 and IMA-3.
| IMA-3 | CMK-1 | CaM | Ca2+ | Kd(mean ± SEM, µM) | R2 fit | p vs. -CaM/-Ca2 control | p vs. CMK-1(wt) |
|---|---|---|---|---|---|---|---|
| + | wt | - | - | 1.54 ± 0.09 | 0.991 | - | - |
| + | wt | + | + | 0.45 ± 0.05 | 0.989 | <0 .0001 | - |
| + | - | + | + | No detectable interaction | - | - | - |
| + | wt | - | + | 1.14 ± 0.19 | 0.970 | ns | - |
| + | K71A/R74Q/R77S | - | - | 1.22 ± 0.09 | 0.988 | - | ns |
| + | K71A/R74Q/R77S | + | + | 1.00 ± 0.15 | 0.963 | ns | <0.05 |
-
ns, not significant.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Caenorhabditis elegans) | N2 | CGC;RRID:SCR_007341 | Wild-type | |
| Genetic reagent (C. elegans) | DAG439 | This study | domSi439[mec-3p::cmk-1 (1–348)::mNG::unc-54 3’UTR] II | Expression of CMK-1(wt)::mNG in FLP |
| Genetic reagent (C. elegans) | PHX1375 | This study | cmk-1(syb1375) IV | Allele encoding CMK-1 (V292A/V294A)Mutation made by genome editing (SunyBiotech, China) |
| Genetic reagent (C. elegans) | PHX1435 | This study | cmk-1(syb1435) IV | Allele encoding CMK-1(R77S)Mutation made by genome editing (SunyBiotech, China) |
| Genetic reagent (C. elegans) | DAG356 | Marques et al., 2019 | domIs272a[mec-3p::QF, mec-4p::QS, QUAS::CoCHR, unc-122p::RFP] | [FLP::CoChR] FLP optogenetic background, used as background for light-avoidance assays |
| Genetic reagent (C. elegans) | DAG821 | This study | cmk-1(ok287) IV | cmk-1 loss-of-function allele4× backcrossed |
| Genetic reagent (C. elegans) | DAG800 | Schild et al., 2014 | cmk-1(pg58) IV | cmk-1 gain-of-function allele4× backcrossed |
| Genetic reagent (C. elegans) | DAG874 | Marques et al., 2019 | unc-68(dom13) V | unc-68 gain-of-function allele4× backcrossed |
| Genetic reagent (C. elegans) | DAG927 | This study | domSi439[mec-3p::cmk-1::mNG::3xFlag::unc-54 3’UTR] II; unc-31(e928) IV | Expression of CMK-1(wt)::mNG in FLP in unc-31 mutant background |
| Genetic reagent (C. elegans) | DAG928 | This study | domSi439[mec-3p::cmk-1::mNG::3xFlag::unc-54 3’UTR] II; unc-13(e450) I | Expression of CMK-1(wt)::mNG in FLP in unc-13 mutant background |
| Genetic reagent (C. elegans) | DAG1032 | This study | domSi439[mec-3p::cmk-1::mNG::3xFlag::unc-54 3’UTR] II;[unc-68(dom13)] V | Expression of CMK-1(wt)::mNG in FLP in unc-68 mutant background |
| Genetic reagent (C. elegans) | DAG1204 | This study | cmk-1(ok287) IV; domIs272[mec-3p::QF, mec-4p::QS, QUAS::CoCHR, unc-122p::RFP] II | cmk-1 loss-of-function allele in [FLP::CoChR] FLP optogenetic background for light-avoidance assays |
| Genetic reagent (C. elegans) | DAG1205 | This study | cmk-1 (syb1375) IV; domIs272[mec-3p::QF, mec-4p::QS, QUAS::CoCHR, unc-122p::RFP] II | cmk-1 genome-edited mutation coding for CMK-1(V292A/V294A) in [FLP::CoChR] FLP optogenetic background for light-avoidance assays |
| Genetic reagent (C. elegans) | DAG1206 | This study | cmk-1(syb1435) IV; domIs272[mec-3p::QF, mec-4p::QS, QUAS::CoCHR, unc-122p::RFP] II | cmk-1 genome-edited mutation coding for CMK-1(R77S) in [FLP::CoChR] FLP optogenetic background for light-avoidance assays |
| Genetic reagent (C. elegans) | GN2 | Gift frm Miriam Goodman | oyIs17[gcy-8p::GFP] | gcy-8 transcriptional reporter |
| Genetic reagent (C. elegans) | VC220 | CGC; RRID:SCR_007341 | cmk-1(ok287); oyIs17[gcy-8p::GFP] | gcy-8 transcriptional reporter in cmk-1 loss-of-function background |
| Genetic reagent (C. elegans) | DAG1053 | This study | cmk-1(syb1375) IV; oyIs17[gcy-8p::GFP] | gcy-8 transcriptional reporter in cmk-1 genome-edited mutant background coding for CMK-1(V292A/V294A) |
| Genetic reagent (C. elegans) | DAG1054 | This study | cmk-1(syb1435) IV; oyIs17[gcy-8p::GFP] | gcy-8 transcriptional reporter in cmk-1 genome-edited mutant background coding for CMK-1(R77S) |
| Genetic reagent (C. elegans) | DAG977 | This study | domSi439[mec-3p::cmk-1::mNG::3xFlag::unc-54 3’UTR] II; osm-9(ky10) ocr-2(ak47) IV | Expression of CMK-1(wt)::mNG in FLP in osm-9;ocr-2 double mutant background |
| Genetic reagent (C. elegans) | DAG978 | This study | domSi439[mec-3p::cmk-1::mNG::3xFlag::unc-54 3’UTR] II; unc-68(r1161) V | Expression of CMK-1(wt)::mNG in FLP in unc-68 loss-of-function mutant background |
| Genetic reagent (C. elegans) | DAG979 | This study | domSi439[mec-3p::cmk-1::mNG::3xFlag::unc-54 3’UTR] II; unc-2(gk366) X | Expression of CMK-1(wt)::mNG in FLP in unc-2 mutant background |
| Genetic reagent (C. elegans) | DAG980 | This study | domSi439[mec-3p::cmk-1::mNG::3xFlag::unc-54 3’UTR] II; unc-2(ra612) X | Expression of CMK-1(wt)::mNG in FLP in unc-2 mutant background |
| Genetic reagent (C. elegans) | DAG981 | This study | domSi439[mec-3p::cmk-1::mNG::3xFlag::unc-54 3’UTR] II; egl-19(n582) IV | Expression of CMK-1(wt)::mNG in FLP in egl-19 mutant background |
| Genetic reagent (C. elegans) | DAG565-566-567 | This study | domEx565-566-567[mec-3p::cmk-1(Δ315–323)::mNG, unc-122p::RFP] | Expression of CMK-1(Δ315–323)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG576-577-578 | This study | domEx576-577-578[mec-3p::cmk-1(Δ318–324)::mNG, unc-122p::RFP] | Expression of CMK-1(Δ318–324)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG592-593 | This study | domEx592-593[mec-3p::CMK-1(Δ288–294)::mNG, unc-122p::RFP] | Expression of CMK-1(Δ288–294)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG703-704-705 | This study | domEx703-704-705[mec-3p::cmk-1(V292A/V294A)::mNG, unc-122p::RFP] | Expression of CMK-1(V292A/V294A)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG706-707-708 | This study | domEx706-707-708[mec-3p::cmk-1(K71A/R74A/R77S)::mNG, unc-122p::RFP] | Expression of CMK-1(K71A/R74A/R77S)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG727-728-729 | This study | domEx727-728-729[mec-3p::cmk-1(L321A/L323A)::mNG, unc-122p::RFP] | Expression of CMK-1 (L321A/L323A)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG1009-1010-1011 | This study | domEx1009-1010-1011[mec-3p::cmk-1(I315A)::mNG, unc-122p::RFP] | Expression of CMK-1(I315A)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG1012-1013-1014 | This study | domEx1012-1013-1014[mec-3p::cmk-1(I315A/L321A/L323A)::mNG, unc-122p::RFP] | Expression of CMK-1(I315A/L321A/L323A)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG738-739-740 | This study | domEx738-739-740[mec-3p::cmk-1(K307Q)::mNG, unc-122p::RFP] | Expression of CMK-1(K307Q)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG741-742-743 | This study | domEx741-742-743[mec-3p::cmk-1(R302S/K307Q)::mNG, unc-122p::RFP] | Expression of CMK-1(R302S/K307Q)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG962-963-966 | This study | domEx962-963-966[mec-3p::cmk-1(K71A)::mNG, unc-122p::RFP] | Expression of CMK-1(K71A)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG964-965 | This study | domEx964-965[mec-3p::cmk-1(R74Q)::mNG, unc-122p::RFP] | Expression of CMK-1(R74Q)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG967-968-969 | This study | domEx967-968-969[mec-3p::cmk-1(R77S)::mNG, unc-122p::RFP] | Expression of CMK-1(R77S)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG900-901-902 | This study | domEx900-901-902[mec-3p::cmk-1(K71A/R74Q/R77S/V292A/V294A)::mNG, unc-122p::RFP] | Expression of CMK-1(K71A/R74Q/R77S/V292A/V294A)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG700-701-702 | This study | domEx700-701-702[mec-3p::cmk-1(W305S)::mNG, unc-122p::RFP] | Expression of CMK-1(W305S)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG912-913-914 | This study | domEx912-913-914[mec-3p::cmk-1(W305S/V292A/V294A)::mNG, unc-122p::RFP] | Expression of CMK-1(W305S/V292A/V294A)::mNG in FLP |
| Genetic reagent (C. elegans) | DAG1029-1030-1031 | This study | domEx1029-1030-1031[ima-3p::egl-13NLS::wrmScarlet::unc-54 3’UTR, unc-122p::GFP]; domSi439[mec-3p::cmk-1::mNG::3xFlag::unc-54 3’UTR] II | Transcriptional reporter for ima-3 driving the expression of a red nuclear marker in a background with FLP labeled in green |
| Genetic reagent (C. elegans) | DAG1244-1245-1246 | This study | domEx1244-1245-1246[mec-3p::cmk-1(R77S)::mNG, unc-122p::RFP];[unc-68(dom13)]V | Expression of CMK-1(R77S)::mNG in FLP in a unc-68 gain-of-function background |
| Genetic reagent (C. elegans) | DAG1279-1280-1281 | This study | domEx1279-1280-1281[mec-3p::cmk-1(K71A/R74Q/R77S)::mNG, unc-122p::RFP];[unc-68(dom13)]V | Expression of CMK-1(K71A/R74Q/R77S)::mNG in FLP in a unc-68 gain-of-function background |
| Genetic reagent (C. elegans) | DAG1282-1283-1284 | This study | domEx1282-1283-1284[mec-3p::cmk-1(W305S)::mNG, unc-122p::RFP];[unc-68(dom13)]V | Expression of CMK-1(W305S)::mNG in FLP in a unc-68 gain-of-function background |
| Genetic reagent (C. elegans) | DAG1436-1437-1438 | This study | domEx1436-1437-1438[cmk-1p::cmk-1(wt)::mNG, unc-122p::GFP]; domIs272[mec-3p::QF, mec-4p::QS, QUAS::CoCHR, unc-122p::RFP] II; cmk-1(syb1375) IV | Rescue of cmk-1(syb1375) using cmk-1 promoter, in the [FLP::CoChR] optogenetic background |
| Genetic reagent (C. elegans) | DAG1439-1440-1441 | This study | domEx1439-1440-1441[mec-3p::cmk-1(wt)::mNG, unc-122p::GFP]; domIs272[mec-3p::QF, mec-4p::QS, QUAS::CoCHR, unc-122p::RFP] II; cmk-1(syb1375) IV | Rescue of cmk-1(syb1375) using mec-3 promoter, in the [FLP::CoChR] optogenetic background |
| Genetic reagent (C. elegans) | DAG1322-1323-1324 | This study | domEx1322-1323-1324[ttx-1p::QF, QUAS::cmk-1(wt)::mNG, unc-122p::RFP] | Expression of CMK-1(wt)::mNG in AFD |
| Genetic reagent (C. elegans) | DAG1325-1326-1327 | This study | domEx1325-1326-1327[ttx-1p::QF, QUAS::cmk-1(K71A/R71Q/R77S)::mNG, unc-122p::RFP] | Expression of CMK-1(K71A/R71Q/R77S)::mNG in AFD |
| Genetic reagent (C. elegans) | DAG1328-1329-1330 | This study | domEx1328-1329-1330[ttx-1p::QF, QUAS::cmk-1(R77S)::mNG, unc-122p::RFP] | Expression of CMK-1(R77S)::mNG in AFD |
| Genetic reagent (C. elegans) | DAG1331-1332-1333 | This study | domEx1331-1332-1333[ttx-1p::QF, QUAS::cmk-1(V292A/V294A)::mNG, unc-122p::RFP] | Expression of CMK-1(V292A/V294A)::mNG in AFD |
| Genetic reagent (C. elegans) | DAG1433-1434-1435 | This study | domEx1433-1434-1435[ttx-1p::QF, QUAS::cmk-1(W305S)::mNG, unc-122p::RFP] | Expression of CMK-1(W305S)::mNG in AFD |
| Genetic reagent (C. elegans) | DAG1469-1470-1471 | This study | domEx1469-1470-1471[ttx-1p::QF, QUAS::cmk-1(wt)::mNG, unc-122p::RFP]; unc-68(dom13) V | Expression of CMK-1(wt)::mNG in AFD in the unc-68 gain-of-function background |
| Genetic reagent (C. elegans) | DAG1472-1473-1474 | This study | domEx1472-1473-1474[ttx-1p::QF, QUAS::cmk-1(R77S)::mNG, unc-122p::RFP]; unc-68(dom13) V | Expression of CMK-1(R77S)::mNG in AFD in the unc-68 gain-of-function background |
| Genetic reagent (C. elegans) | DAG1475-1476-1477 | This study | domEx1475-1476-1477[ttx-1p::QF, QUAS::cmk-1(W305S)::mNG, unc-122p::RFP]; unc-68(dom13) V | Expression of CMK-1(W305S)::mNG in AFD in the unc-68 gain-of-function background |
| Genetic reagent (C. elegans) | DAG1513 | This study | domEx1513[ttx-1prom::QF, QUAS::YC2.3] | Cameleon (YC2.3) expression in AFD for calcium imaging |
| Genetic reagent (C. elegans) | DAG616 | This study | domEx616[mec-3p::calbindin, unc-122p::GFP]; domSi437[mec-3p::cmk-1::mNG::3xFlag::unc-543’UTR] II | Calcium buffering in FLP via the expression of Calbindin |
| Genetic reagent (C. elegans) | AQ2145 | Gift from Bill Schafer | ljEx19[egl-46p::YC2.3; lin15(+)] | Cameleon (YC2.3) expression. |
| Genetic reagent (C. elegans) | DAG747 | Saro et al., 2020 | unc-68(r1161) V; ljEx19[egl-46p::YC2.3; lin15(+)] | Cameleon (YC2.3) expression in FLP for calcium imaging in an unc-68 loos-of-function mutant background. |
| Genetic reagent (C. elegans) | DAG792 | Saro et al., 2020 | egl-19(n582) IV; ljEx19[egl-46p::YC2.3; lin15(+)] | Cameleon (YC2.3) expression in FLP for calcium imaging in egl-19(n582) IV mutant background |
| Genetic reagent (C. elegans) | DAG918 | Saro et al., 2020 | ocr-2(ak47) IV; ljEx19[egl-46p::YC2.3; lin15(+)] | Cameleon (YC2.3) expression in FLP for calcium imaging in ocr-2(ak47) IV mutant background |
| Genetic reagent (C. elegans) | DAG1001 | Saro et al., 2020 | unc-2(ra612) X; ljEx19[egl-46p::YC2.3; lin15(+)] | Cameleon (YC2.3) expression in FLP for calcium imaging in unc-2(ra612) X mutant background |
| Genetic reagent (C. elegans) | DAG857-859 | This study | domSi439[mec-3p::cmk-1::mNeonGreen::3xFlag::unc-54UTR] II; domEx857-859[mec-3p::egl-13NLS::CeBFP::unc-54UTR] | Expression of a blue nuclear marker in FLP already expressing CMK-1::mNG green |
| Genetic reagent (C. elegans) | DAG933-934 | This study | domEx933-934[mec-3p::QF]; [QUAS::mNeonGreen, unc-122p::RFP]; [mec-3p::egl-13NLS_CeBFP::unc-54UTR] | Expression of a blue nuclear marker in FLP already labeled in green |
| Genetic reagent (C. elegans) | DAG1650 | This study | domEx1650[mec-3p::Calb28K::unc54UTR]; [mec-3p::QF::UTR54]; [QUASp::YC2.3::UTR54] | Cameleon (YC2.3) expression in FLP for calcium imaging in presence of Calbindin |
| Genetic reagent (C. elegans) | DAG1652-1653-1654 | This study | domEx1652-1653-1654[mec-3p::rParv::unc-54UTR]; domSi439[mec-3p::cmk-1::mNeonGreen::3xFlag::unc-54UTR] II | Calcium buffering in FLP via the expression of Parvalbumin |
| Genetic reagent (C. elegans) | DAG1655-1656-1657 | This study | domEx1655-1656-1657[mec-3p::rParv_mutant::unc-54UTR]; domSi439[mec-3p::cmk-1::mNeonGreen::3xFlag::unc-54UTR] II | Negative control for calcium buffering in FLP via the expression of mutant Parvalbumin (K92V, D93A, D95A, K97V, and E100V) |
Plasmids and primers used for cloning.
| Plasmid types | Plasmid name, cloning, and primer information |
|---|---|
| Promoter plasmids (Multi-Site Gateway slot 1): | dg701[slot1 Entry ima-3p]attB4ima-3_F: ggggacaagtttgtacaaaaaagcaggcttacatatgagttcaaacagacaggcttattattB1ima-3_R: ggggaccactttgtacaagaaagctgggtcggatccctttccaaagttccatcctccgdg508[slot1 Entry ttx-1p]attB4_F: ggggacaactttgtatagaaaagttgatccatactcaggggaacagtgtattB1_R: ggggactgcttttttgtacaaacttgtgaagcaggaatatatgacaaatgaaatacgThe generation of dg68[slot1 Entry mec-3p(noATG)] and dg229[slot1 Entry QUASprom] was previously described in Schild and Glauser, 2015 and the generation of mg268[slot1 Entry cmk-1p] in Schild et al., 2014. |
| Coding sequence plasmids (Multi-Site Gateway slot 2): | dg703[slot2 Entry ima-3cds]attB1ima3cds_F: ggggacaagtttgtacaaaaaagcaggcttacatatgagttcaaacagacaggcttattattB2ima3cds_R: ggggaccactttgtacaagaaagctgggtcggatccctttccaaagttccatcctccgThe generation of dg240 [slot2 Entry QF_withATG] and dg245[mec-4p(2kb)::QS::SL2mCherry] was previously described in Schild and Glauser, 2015; the generation of dg286 [slot2 Entry (cmk-1cds_noSTOP)] was described in Hostettler et al., 2017; the generation of dg651 [slot2 Entry egl-13NLS::wrmScarlet] was described in Marques et al., 2019; the generation of dg718[slot2 Entry YC2.3] was described in Saro et al., 2020.dg215[pENTR slot2_calb28K] created by PCR amplification from [odr-3p::calbindin D28K] (gift from Chiou-Fen Chuang) followed by subcloning into pDON221 via BP recombination.dg217[pENTR slot2_rParv] created by PCR amplification from [odr-3p:: rParv] (gift from Chiou-Fen Chuang) followed by subcloning into pDON221 via BP recombination.dg219[pENTR slot2_rParv_mutant] created by PCR amplification from [odr-3p:: rParvCDEF/AV] (gift from Chiou-Fen Chuang) followed by subcloning into pDON221 via BP recombination.dg643[pENTR slot2 Ex13_CeBFP-reporter_V2] was created amplifying by PCR and subcloning into pDON221 by BP recombination a codon-optimized CeBFP version containing three artificial introns obtained via gene synthesis (Eurofins DNA).dg650[slot2 Entry egl-13NLS::wrmScarlet] was created adding to dg643 the egl-13NLS by PCR with the following primers:NLSRightHalf_CeBFP_F: aaacgcgaagaagcttgccaaggaagttgaaaatggatccatgtcagagcttattaaggNLSLeftHalf_CeBFP_R: cactcagttttgtcggattcgcttttcgtctacggctcatgttgctagcggtacctaag |
| Plasmids carrying cmk-1 coding sequence with small deletions and primers used to introduce these deletions: | dg168[slot2 Entry cmk-1(Δ315–323)]cmk-1_324F: tcctcaaatagcaatcgcctacagaaaccmk-1_314R: tgctgcggggctgcgttdg173[slot2 Entry cmk-1(Δ318–324)]cmk-1_325F: tcaaatagcaatcgcctacagaaacaagcmk-1_317R: ctggcggattgctgcggdg192[slot2 Entry cmk-1(Δ288–294)]CMK-1_287R: atcgtgtgtgtacgccgtatttcCMK-1_295F: catcttaagaagagtttggcaaaacgga |
| Plasmids with cmk-1 point mutations and primers used for site-directed mutagenesis: | dg588[slot2 Entry cmk-1(K71A/R74Q/R77S)]NLS71-78_F: cagaagctctcacacaacaatattgttcaactattcgaNLS71-78_R: cagaactgcaatctcgttttccagtgattcttcdg589[slot2 Entry cmk-1(W305S)]W305S_F: ccaaaaaggcttacaacgcagccW305S_R: agttccgttttgccaaactcttctdg590[slot2 Entry cmk-1(V292A/V294A)]NES288-294_F: cgctcatcttaagaagagtttggcaaaacggaNES288-294_R: gcggcagttccgtgaatatcgtgtgtgtacdg591[slot2 Entry cmk-1(L321A/L323A)]NES314-323_F: gctcgtgcttcctcaaatagcaatcgcctacagaaNES314-323_R: catttgaagctggcggattgctdg593[slot2 Entry cmk-1(K307Q)]NLS297-308_F: caggcttacaacgcagccgcNLS297-308_R: tttccagttccgttttgccaaactcttcdg612[slot2 Entry cmk-1(R303S/K307Q)]NLS297-308_F: tccaactggaaacaggcttacaacgNLS297-308_R: ttttgccaaactcttcttaagatgtacgdg658[slot2 Entry cmk-1(K71A/R74Q/R77S/V292A/V294A)]NLS71-78_F/R with dg608 as templatedg663[slot2 Entry cmk-1(V292A/V294A/W305S)]W305S_F/R with dg608 as templatedg674[slot2 Entry cmk-1(K71A)]NLS71-78_ K71A_F: aggaagctccgacacaacNLS71-78_ K71A_R: cagaactgcaatctcgttttccagtgattcttcdg675[slot2 Entry cmk-1(R74Q)]NLS71-78_ R74Q_F: cagaagctccgacacaacaatattgttcaactaNLS71-78_ R74Q_R: cagaactttaatctcgttttccagtgattcttcdg676[slot2 Entry cmk-1(R77S)]NLS71-78_ R77S_F: tcacacaacaatattgttcaactattcgaNLS71-78_ R77S_R: gagcttcctcagaactttaatctcdg688[slot2 Entry cmk-1(I315A)]NES314-323_315F: ctccgtctctcctcaaatagcaatcgcNES314-323_315R: catttgaagctggcgggctgctgcgdg689[slot2 Entry cmk-1(I315A/L321A/L323A)]NES314-323_315F/R with dg608 as template |
| 3′ UTR and tagging plasmids (Multi-site Gateway slot 3): | mg277[slot3 Entry SL2::mCherry] was previously described in Schild et al., 2014.mg211[slot3 Entry unc-54 3’UTR] (aka pMH473) was a gift from Marc Hammarlund.dg397[slot3 Entry mNG::3xFLAG::unc-54 3’UTR] previously described in Hostettler et al., 2017. |
| Selection markers used for transgenesis: | dg9 [coel::RFP] (or unc-122p::RFP) was a gift from Piali Sengupta (Addgene plasmid # 8938).dg396 [coel::GFP] (or unc-122p::GFP) was a gift from Piali Sengupta (Addgene plasmid # 8937). |
| Expression plasmids used for transgenesis with a description of their creation: | dg405[mec-3p::cmk-1::mNG::3xFlag] was previously created through an LR recombination reaction between dg68, dg286, dg397 and PCFJ150 (Hostettler et al., 2017).dg425[cmk-1p::cmk-1(wt)::mNG::3xFlag::unc-54 3’UTR] was previously created through an LR recombination reaction between mg268, dg286, dg397, and pDEST-R4-P3 (Hostettler et al., 2017).dg515[mec-3p::cmk-1(Δ315–323)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg168, dg397, and pDEST-R4-P3.dg520[mec-3p::cmk-1(Δ318–324)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg173, dg397, and pDEST-R4-P3.dg524[mec-3p::cmk-1(Δ288–294)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg192, dg397, and pDEST-R4-P3.dg605[mec-3p::cmk-1(K71A/R74Q/R77S)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg588, dg397, and pDEST-R4-P3.dg606[mec-3p::cmk-1(W305S)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg589, dg397, and pDEST-R4-P3.dg607[mec-3p::cmk-1(V292A/V294A)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg590, dg397, and pDEST-R4-P3.dg608[mec-3p::cmk-1(L321A/L323A)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg591, dg397, and pDEST-R4-P3.dg610[mec-3p::cmk-1(K307Q)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg593, dg397, and pDEST-R4-P3.dg613[mec-3p::cmk-1(R303S/K307Q)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg612, dg397, and pDEST-R4-P3.dg665[mec-3p::cmk-1(K71A/R74Q/R77S/V292A/V294A)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg658, dg397, and pDEST-R4-P3.dg670[mec-3p::cmk-1(V292A/V294A/W305S)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg663, dg397, and pDEST-R4-P3.dg677[mec-3p::cmk-1(K71A)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg674, dg397, and pDEST-R4-P3.dg678[mec-3p::cmk-1(R74Q)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg675, dg397, and pDEST-R4-P3.dg679[mec-3p::cmk-1(R77S)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg676, dg397, and pDEST-R4-P3.dg694[mec-3p::cmk-1(I315A)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg688, dg397, and pDEST-R4-P3.dg695[mec-3p::cmk-1(I315A/L321A/L323A)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg689, dg397, and pDEST-R4-P3.dg705[ima-3p::egl-13NLS::wrmScarlet::unc-54 3’UTR] was created through an LR recombination reaction between dg701, dg651, mg211, and pDEST-R4-P3.dg876[QUAS::cmk-1(wt)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg229, dg286, dg397, and pDEST-R4-P3.dg877[QUAS::cmk-1(K71A/R74Q/R77S)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg229, dg588, dg397, and pDEST-R4-P3.dg878[QUAS::cmk-1(R77S)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg229, dg286, dg676, and pDEST-R4-P3.dg879[QUAS::cmk-1(V292A/V294)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg229, dg590, dg397, and pDEST-R4-P3.dg882[QUAS::cmk-1(W305S)::mNG::3xFlag::unc-54 3’UTR] was created through an LR recombination reaction between dg229, dg589, dg397, and pDEST-R4-P3.dg883[ttx-1p::QF::unc-54 3’UTR] was created through an LR recombination reaction between dg508, dg240, mg211, and pDEST-R4-P3.dg777[QUAS::YC2.3::unc-54 3’UTR] was created through an LR recombination reaction between dg229, dg718, mg211 , and pDEST-R4-P3.dg280[mec-3p::Calb28K::unc54UTR] was created through an LR recombination reaction between dg68, dg215, mg211, and pDEST-R4-P3dg282[mec-3p::rParv::unc-54UTR] was created through an LR recombination reaction between dg68, dg217, mg211, and pDEST-R4-P3.dg284[mec-3p::rParv_mutant::unc-54UTR] was created through an LR recombination reaction between dg68, dg219, mg211, and pDEST-R4-P3.dg373[QUAS::NeonGreen] was created through an LR recombination reaction between dg229 (pENTR_slot1_QUASprom) dg353, mg211, and pDEST-R4-P3.dg653[mec-3p::egl-13NLS::CeBFP::unc-54 3’UTR] was created through an LR recombination reaction between dg68, dg650, mg211, and pDEST-R4-P3. |
| Plasmids used for recombinant protein expression: | dg918[ima-3::His-6] was created by inserting ima-3 coding sequence using NdeI and BamHI restriction sites in pDK2832 (Gift from Dieter Kressler). The ima-3 coding sequence was obtained from a modified dg703 plasmid in which an internal BamHI site had been removed using the following primers:Ima-3Bamless_F: cgatccaaacttgcaatttgaagctgIma-3Bamless_R: gtgctggacaagcattgaacgdg922 [cmk-1(wt)::GST-TEV] was created by insertion of cmk-1 cds using NdeI and BamHI restriction sites in pDK2409 (Gift from Dieter Kressler).dg929[cmk-1(K71A/R74Q/R77S)::GST-TEV] was created by PCR site-directed mutagenesis from dg922. |





