Differences in local immune cell landscape between Q fever and atherosclerotic abdominal aortic aneurysms identified by multiplex immunohistochemistry
Figures
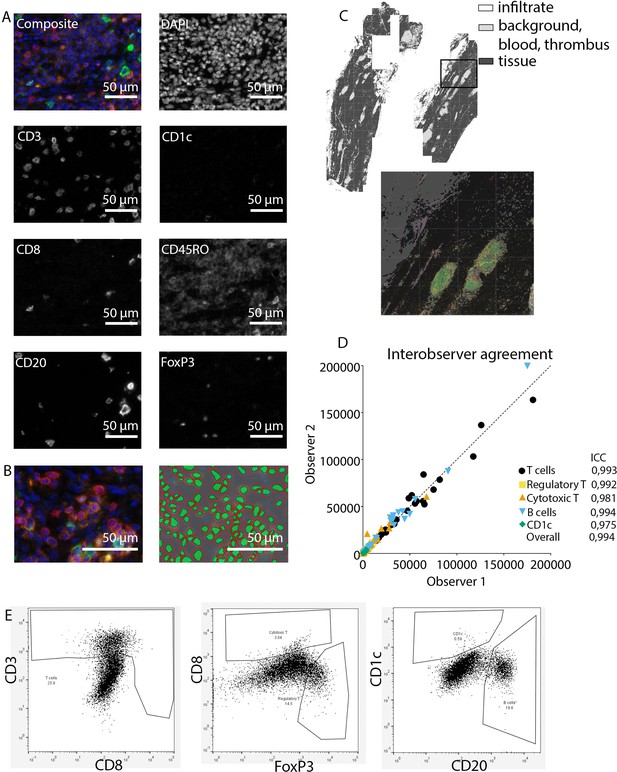
Analysis of adaptive immune system panel.
(A) Composite image and separate channels. (B) Composite image and cell segmentation tool. (C) Tissue segmentation tool (upper image) and overlay with staining, which shows overlay in segmented and actual stained infiltrate. (D) Interobserver agreement with high intraclass coefficients, supporting our robust method. (E) Representative FACS plots for drawing cell populations.
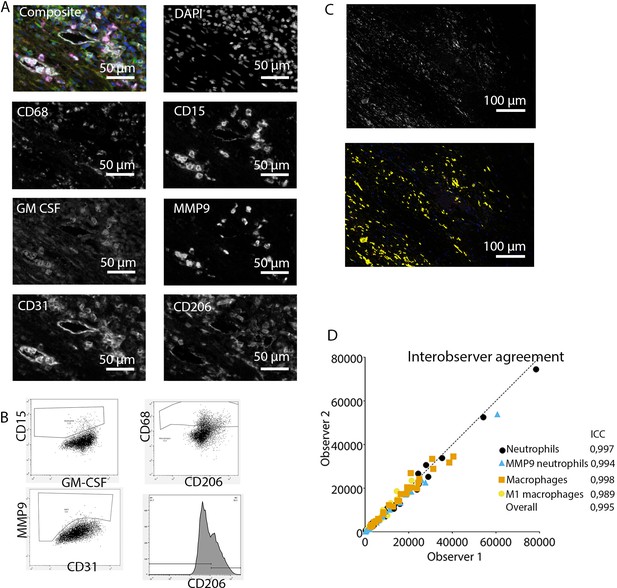
Analysis of innate immune system panel.
(A) Composite image and separate channels. (B) Representative FACS plots for drawing cell populations. (C) inForm threshold analysis of GM-CSF expression. Upper image: GM-CSF channel. Lower image: GM-CSF signal above threshold. (D) Interobserver agreement with high intraclass coefficients.
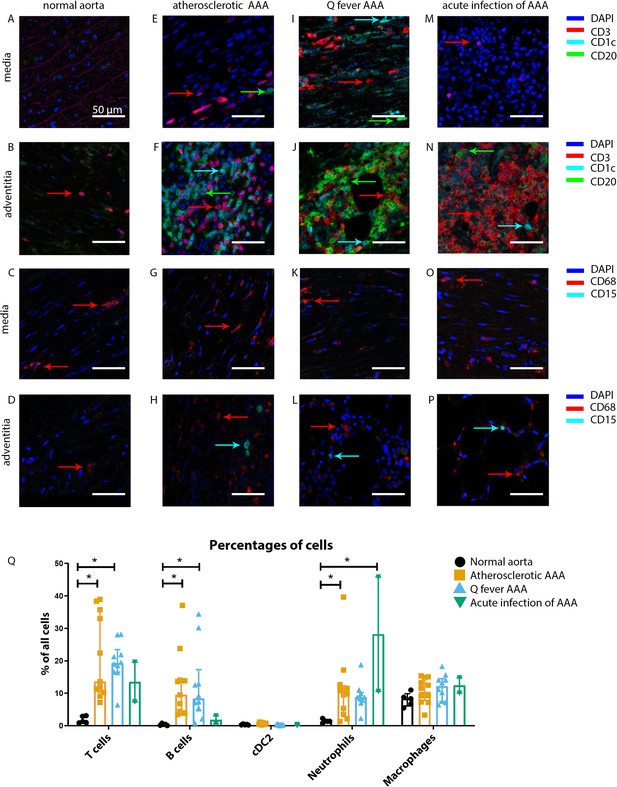
Immune cell activation in atherosclerotic, Q fever infected, and acutely infected AAAs.
All scale bars represent 50 µm. (A–P): Adaptive (A, B, E, F, I, J, M, N) and innate (C, D, G, H, K, L, O, P) immune cells in a representative normal abdominal aorta, atherosclerotic AAA, Q fever AAA, and acutely infected AAA. Arrows with corresponding colors indicate the presence of immune cells with red for CD3+ T cells, cyan for CD1c+ cDC2, and green for CD20+ B cells in the adaptive panel (A, B, E, F, I, J, M, N); and red for CD68+ macrophages and cyan for CD15+ neutrophils in the innate panel (C, D, G, H, K, L, O, P). (Q): quantification of percentages of different types of immune cells in the whole tissue sections, showing the increases in T and B cells in atherosclerotic AAA and Q fever AAA compared to normal and increase in neutrophils in acute infection and atherosclerotic AAA compared to normal. Note that there are no differences between atherosclerotic AAA and Q fever AAA. * Represents p≤0.05. Source data can be found in Figure 3—source data 1.
-
Figure 3—source data 1
Immune cell activation in atherosclerotic, Q fever infected, and acutely infected AAAs.
- https://cdn.elifesciences.org/articles/72486/elife-72486-fig3-data1-v2.xlsx
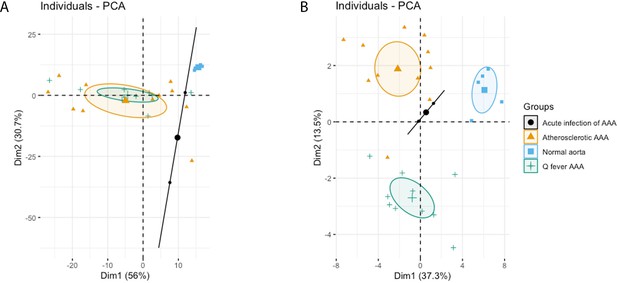
Principal Component Analyses.
(A) Principal component analysis (PCA) including CD3, CD20, CD68, CD15, and CD1c. There is a clear distinct population consisting of normal abdominal aortas. There are two data points for acute infection, resulting in a line. Intriguingly, atherosclerotic AAA and Q fever infected AAA are completely overlapping. This indicates that these populations are similar when testing for these cell markers. (B): PCA including all markers (CD68, CD15, MMP9, GMCSF, CD31, CD206, CD3, CD1c, CD8, FoxP3, CD45RO, and CD20). Note the difference with (A): here all groups form separate populations, indicating that the newly added markers including subset markers describe the differences between atherosclerotic and Q fever AAA. See Supplementary file 1A for loadings of both PCAs.
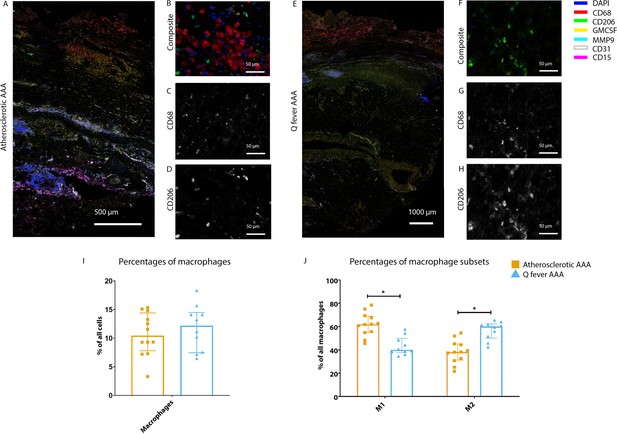
Phenotype shift in macrophages in Q fever AAA towards M2.
(A): Overview photo of atherosclerotic AAA, upper portion is intima layer, lower portion adventitia. (B): Composite of CD68 and CD206 with majority CD68. (C, D): Separated channels for CD68 and CD206, respectively. (E): Overview of Q fever infected AAA, with the same orientation as (A). (F): Composite of CD68 and CD206, with mostly CD206+ cells which also express CD68, as supported by separated channels in (G) and (H). (I, J): Quantification of percentages of macrophages in entire tissue sections (I) and of proportions of M1 and M2 macrophages in these macrophages (J), showing the phenotype switch in Q fever AAAs toward M2. * Represents p≤0.05. Source data can be found in Figure 5—source data 1.
-
Figure 5—source data 1
Phenotype shift in macrophages in Q fever AAA towards M2.
- https://cdn.elifesciences.org/articles/72486/elife-72486-fig5-data1-v2.xlsx
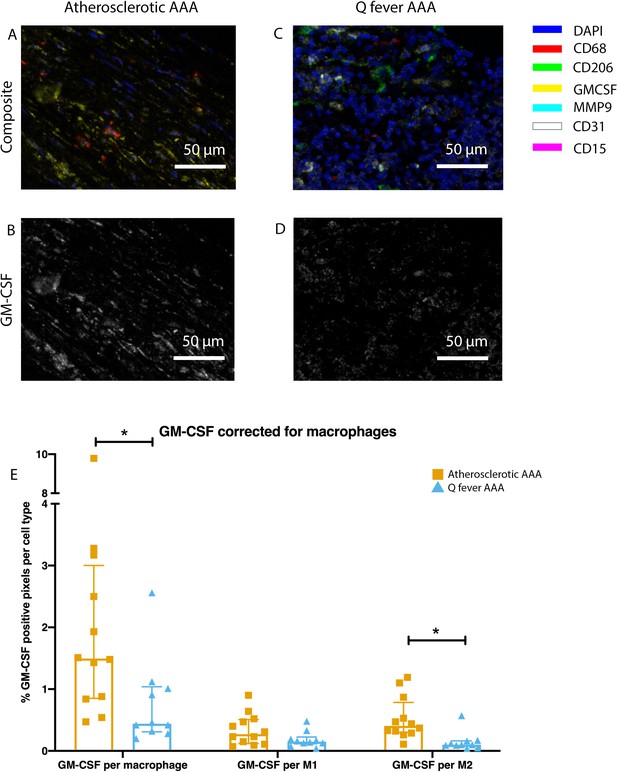
Q fever infected AAAs express lower levels of GM-CSF.
(A–D): Representative composite image of atherosclerotic AAA (A) and Q fever AAA (C) and corresponding GM-CSF channels (B, D). (E): The expressed levels of GM-CSF corrected for the number of macrophages and M2 macrophages are lower in Q fever infected AAAs, suggesting an immune-suppressed environment. Source data can be found in Figure 6—source data 1.
-
Figure 6—source data 1
Q fever infected AAAs express lower levels of GM-CSF.
- https://cdn.elifesciences.org/articles/72486/elife-72486-fig6-data1-v2.xlsx
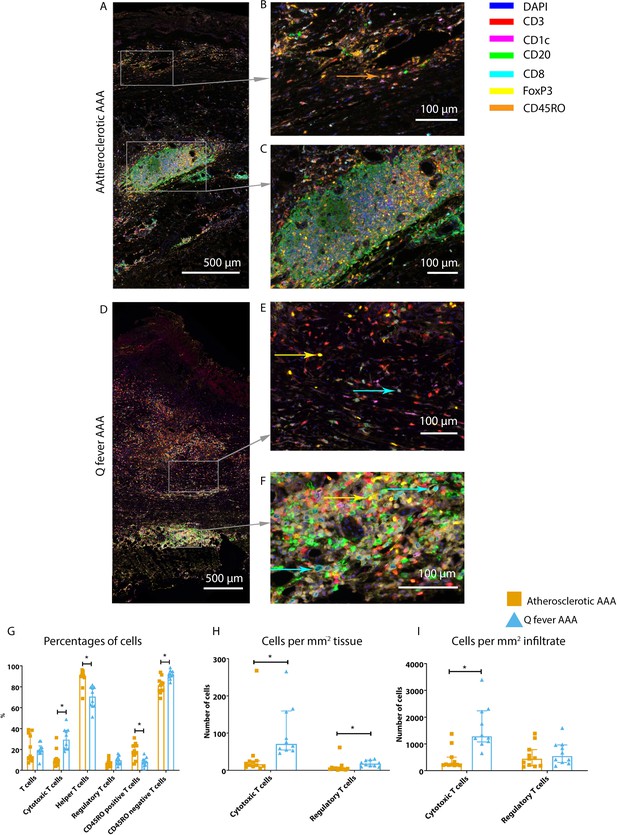
Q fever AAAs exhibit both pro-inflammatory and anti-inflammatory T cell subsets.
Arrows with corresponding colors indicate the presence of immune cells, with orange for memory T cells, yellow for T helper cells, and cyan for cytotoxic T cells. (A–F): Overview of atherosclerotic AAA (A) and Q fever AAA with zoomed photos of tissue (B, E) and tertiary lymphoid structures (TLS) (C, F). In both (A) and (B), the upper side of the photo is the intima layer. Note all the FoxP3+ (yellow) cells in Q fever infected tissue. (G): Percentage of T cells of all cells and T cells subsets out of T cells; (G, H, I): Quantification shows a shift in cytotoxic/helper T cell ratio and decrease in memory T cells in Q fever AAAs. Q fever AAAs show increased numbers of cytotoxic and regulatory T cells, indicating both immune activation and suppression. Source data can be found in Figure 7—source data 1.
-
Figure 7—source data 1
Q fever AAAs exhibit both pro-inflammatory and anti-inflammatory T cell subsets.
- https://cdn.elifesciences.org/articles/72486/elife-72486-fig7-data1-v2.xlsx
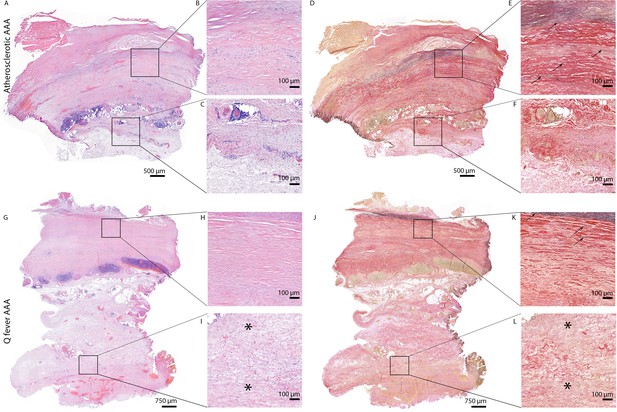
HE and Elastin von Gieson (EVG) stainings demonstrate the disrupted architecture of Q fever infected AAAs.
Representative images of HE staining of AAA with 22× zoomed-in sections (A–C) and EVG staining of adjacent slide (D–F) demonstrate the atherosclerotic plaque, immune cells, and infiltrates with relatively preserved vessel architecture as shown by presence of elastin fibers (black arrows pointing at black lines). (HE) (G–I) and EVG (J–L) of adjacent Q fever AAAs slides reveal pronounced atherosclerosis and immune cell infiltration, and loss of elastin fibers in the media layer (K). In the adventitia (I, L), tissue is replaced by large amounts of fibrosis, indicated with asterisks.
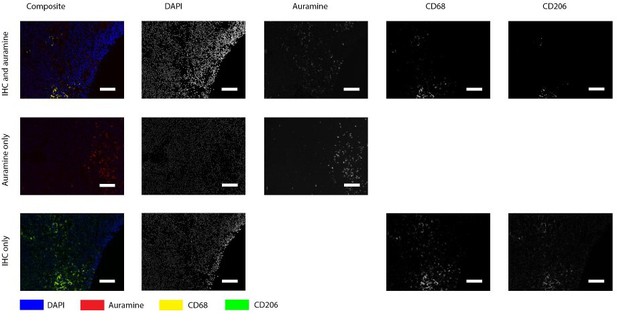
Positive control for Auramin staining.
Tissue was stained with both IHC and Auramine (first row), only with Auramine (middle row), or scanned between the IHC and Auramine steps (last row). Auramine channel shows numerous positive areas in the infected tissue (goat lymph node). All scale bars represent 50 µm.
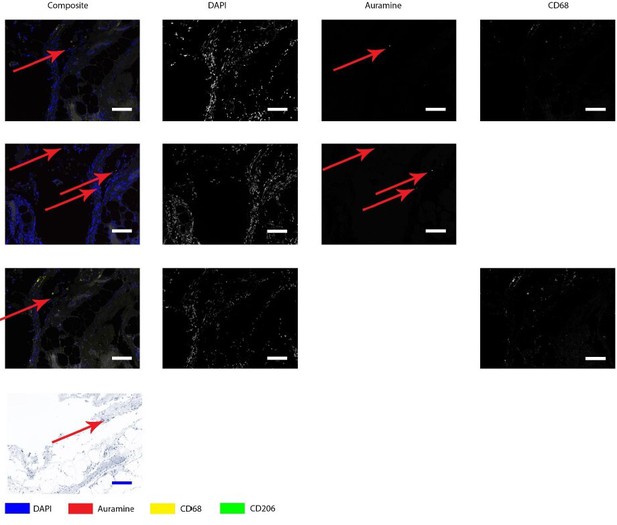
Auramine and Kinyoun staining on Q fever infected AAA, magnification in adventitia.
Tissue was stained with both IHC and Auramine (first row), only with Auramine (middle row), or scanned between the IHC and Auramine steps (last row). Some Auramine-positive spots are present (red arrows). In the IHC only and Kinyoun images these arrows point at corresponding locations. In Kinyoun staining no bacterium was found. All scale bars represent 50 µm..
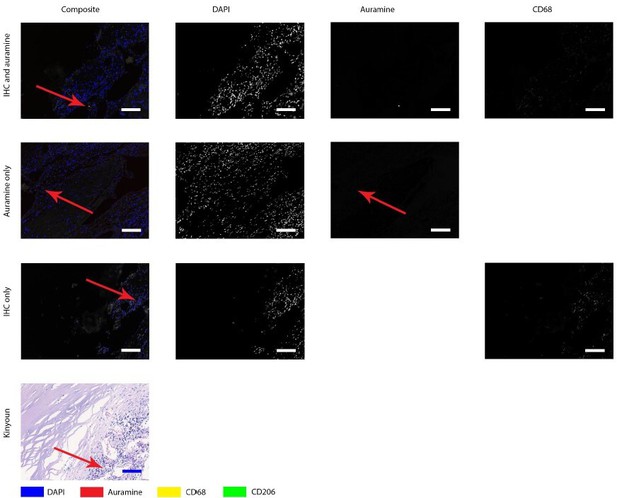
Auramine and Kinyoun staining on Q fever infected AAA, magnification in intima.
Tissue was stained with both IHC and Auramine (first row), only with Auramine (middle row), or scanned between the IHC and Auramine steps (last row). Some Auramine-positive spots are present (red arrows). In the IHC only and Kinyoun images these arrows point at corresponding locations. In Kinyoun staining no bacterium was found. All scale bars represent 50 µm..
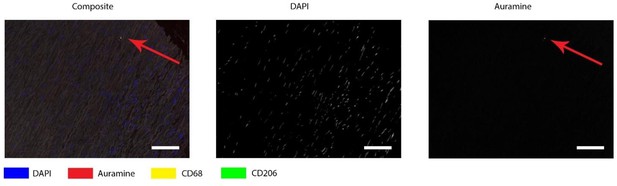
Auramine staining on normal aorta, magnification in media.
In media an Auramine-positive structure was found, in granular shape similar to described in earlier figures. All scale bars represent 50 μm.
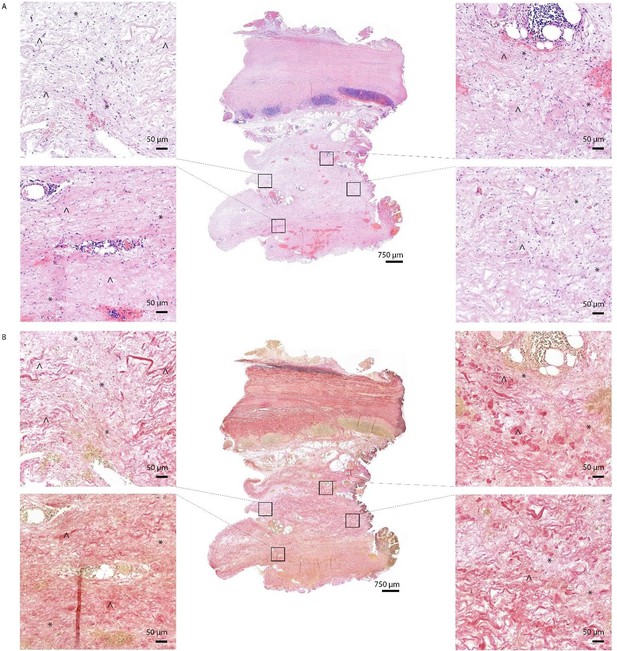
HE (A) and EVG (B) staining of Q fever AAA demonstrate damaged vessel wall architecture.
EVG stained images (B) demonstrate fine fibrotic areas (marked with *) enclosed by pre-existent adventitial structures (marked with ^), mainly consisting of collagen bundles and smooth muscle cells.
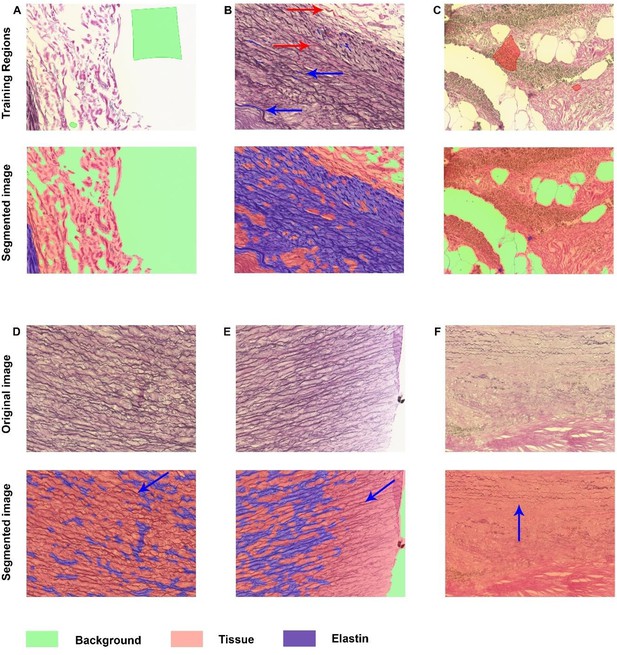
Elastin analysis in inForm software.
A-C: Representative images of training regions (drawn colored regions in green, blue, and red), which are very small in B and therefore indicated with arrows with corresponding colors. The segmented images show nice segmentation of background, tissue and elastin. D-F: These images demonstrate the imperfections of the algorithm: not all elastin fibers are recognized as indicated with blue arrows.
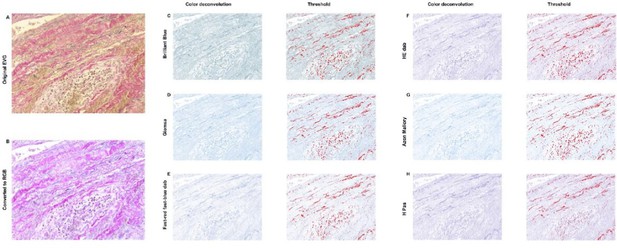
Elastin analysis in FIJI.
The original stained image (A) was converted into RGB (B). C-H: Color deconvolution was performed, which resulted in these segmented images with corresponding thresholds. For all options it was not possible to exclude cells in this threshold while including elastin, resulting in an unreliable analysis.
Tables
Overview of the used markers and clones per panel, including definition of each cell type as used for our analysis.
| Adaptive immune system | Innate immune system | |||
|---|---|---|---|---|
| Markers (clone) | DAPI | DAPI | ||
| CD3 (SP7) | CD68 (PG-M1) | |||
| CD8 (CD8/144B) | CD206 (CL038+) | |||
| CD20 (L26) | CD15 (MMA) | |||
| CD1c (2F4) | CD31 (JC70A) | |||
| FoxP3 (236A/E7) | MMP9 (polyclonal) | |||
| CD45RO (UCHL-1) | GM-CSF (polyclonal) | |||
| Autofluorescence | Autofluorescence | |||
| Cell phenotype | T cell | CD3+ | Macrophage | CD68+ |
| Helper T cell | CD3+ CD8− | M1-like macrophage | CD68+ CD206− | |
| Cytotoxic T cell | CD3+ CD8+ | M2-like macrophage | CD68+ CD206+ | |
| Regulatory T cell | CD3+ CD8− FoxP3+ | Neutrophil | CD15+ | |
| Memory T cell | CD3+ CD45RO+ | Endothelium | CD31+ | |
| B cell | CD20+ | MMP9+ cell | MMP9+ CD15− | |
| Classic DC type 2 | CD1c+ CD20− | MMP9+ neutrophil | MMP9+ CD15+ | |
| DAPI | Nucleus | DAPI | Nucleus | |
| Autofluorescence | Elastin fibers | Autofluorescence | Elastin fibers | |
Baseline characteristics.
| Characteristic | Normal (N=5) | Missing data normal | Atherosclerotic AAA (N=12) | Missing data atherosclerotic | Q fever AAA (N=10) | Missing data Q fever | Infectious AAA (N=2) | Missing data infectious | Significance |
|---|---|---|---|---|---|---|---|---|---|
| Male | 3 (60%) | 2 | 10 (83.3%) | 0 | 9 (90.0%) | 1 | 1 (50.0%) | 1 | 0.482 |
| Age | 65 (54–68) | 0 | 72 (66–78) | 0 | 71 (64–77) | 0 | 67 (62–) | 0 | 0.307 |
| Length | 1.80 (1.69–1.83) | 0 | 1.77 (1.69–1.80) | 0 | 1.78 (1.70–) | 8 | 1.76–(1.76–1.76) | 1 | 0.923 |
| Weight | 80.0 (77.5–102.5) | 0 | 82.6 (75.5–92.6) | 0 | 88.5 (79–) | 8 | 94.8 (94.8–94.8) | 1 | 0.637 |
| BMI | 27.0 (22.1–34.7) | 0 | 26.8 (24.0–29.4) | 0 | 27.8 (27.3–) | 8 | 30.6 (30.6–30.6) | 1 | 0.651 |
| Hypertension | 3 (60%) | 0 | 8 (66.7%) | 0 | 6 (60.0%) | 2 | 1 (50.0%) | 1 | 1.000 |
| Hypercholesterolemia | 5 | 9 (75.0%) | 0 | 6 (60.0%) | 2 | 0 (0.0%) | 1 | 0.426 | |
| DM | 1 (20%) | 0 | 3 (25.0%) | 0 | 1 (10.0%) | 2 | 0 (0.0%) | 1 | 0.853 |
| Total cholesterol | 5 | 3.6 (3.0–5.2) | 5 | 4.7 (3.2–6.1) | 5 | 2 | 0.371 | ||
| HDL | 5 | 0.9 (0.8–1.1) | 6 | 0.9 (0.9–1.3) | 5 | 2 | 0.583 | ||
| Previous aorta surgery | 0 (0.0%) | 0 | 6 (50.0%) | 0 | 0 (0.0%) | 0 (0.0%) | 1 | 0.034* | |
| Rupture | 0 (0.0%) | 0 | 0 (0.0%) | 0 | 3 (30.0%) | 0 | 0 (0.0%) | 0 | 0.193 |
| Artery disease | 1 (20%) | 4 | 11 (91.7%) | 0 | 4 (40.0%) | 2 | 1 (50.0%) | 1 | 0.002* |
| Smoking | 3 (60%) | 5 | 11 (91.7%) | 1 | 6 (60.0%) | 1 | 1 (50.0%) | 1 | 0.387 |
| Packyears | 27 (27–27) | 4 | 33 (20–50) | 2 | 45 (0–53) | 1 | 0 (0–0) | 1 | 0.819 |
| CRP | 5 | 12 | 12.5 (12.0–65) | 4 | 2 | n/a | |||
| Diameter CT | 5 | 57 (55–71) | 0 | 60 (45–81) | 1 | 46 (46–46) | 1 | 0.397 | |
| Beta blocker | 1 (20%) | 3 | 8 (66.7%) | 0 | 3 (30.0%) | 2 | 0 (0.0%) | 1 | 0.567 |
| ARB/ACEi | 2 (40%) | 2 | 6 (50.0%) | 0 | 3 (30.0%) | 2 | 0 (0.0%) | 1 | 0.789 |
| Calciumblocker | 0 (0%) | 3 | 3 (25.0%) | 0 | 0 (0.0%) | 2 | 0 (0.0%) | 1 | 0.439 |
| Diuretics | 0 (0%) | 4 | 1 (8.3%) | 0 | 2 (20.0%) | 0 | 1 (50.0%) | 1 | 0.009* |
-
* Represents p≤0.05. Numbers display numbers of patients with percentage or median with interquartile range (IQR).
Additional files
-
Supplementary file 1
Loadings of Principal Component Analysis and Overview of Reagents and dilutions.
- https://cdn.elifesciences.org/articles/72486/elife-72486-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/72486/elife-72486-transrepform1-v2.docx





