A genetic compensatory mechanism regulated by Jun and Mef2d modulates the expression of distinct class IIa Hdacs to ensure peripheral nerve myelination and repair
Figures
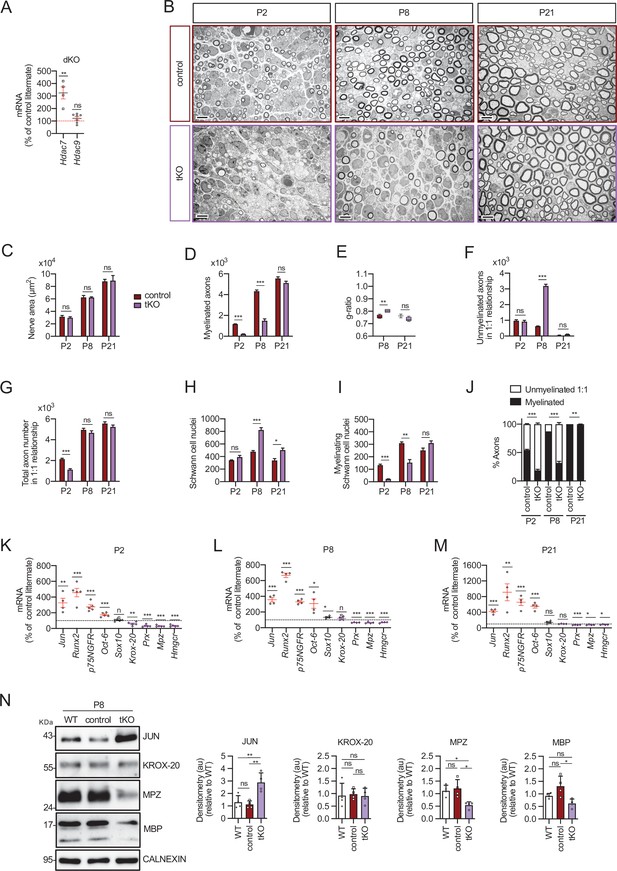
Myelin development is notably delayed in the tKO mice.
(A) A 325.1 ± 48.1% (p = 0.0034) increase in the amount of mRNA for Hdac7 was found in the dKO nerves. No changes in the expression of HDAC9 were found. RT-qPCR with mouse-specific primers for Hdac7 was performed and normalized to 18S rRNA. The scatter plot, which include also the mean ± standard error (SE), shows the fold change in mRNA normalized to control littermates. Four to eight mice per genotype were used. Data were analyzed with the unpaired t-test. (B) Representative transmission TEM mages of P2, P8, and P21 sciatic nerves of tKO mice (Mpz-Cre+/−;Hdac4flx/flx;Hdac5−/−;Hdac7flx/flx) and the control (Mpz-Cre−/−;Hdac4flx/flx; Hdac5−/−;Hdac7flx/flx) littermates. Scale bar: 5 μm. (C) No statistically significant differences were observed between the area of the tKO nerves and control littermates (P2: p = 0.5234; P8: p = 0.9279; P21: p = 0.9009). (D) The number of myelinated axons is notably decreased at P2 (208 ± 24 in tKO versus 1.160 ± 29 in controls; p ≤ 0.0001) and P8 (1.487 ± 179 in tKO versus 4.235 ± 129 in controls; p ≤ 0.0001). (E) g ratio was increased at P8 (0.80 ± 0.01 in the tKO versus 0.76 ± 0.01 in control; p = 0.0045). (F) The number of unmyelinated axons in a 1:1 relationship with Schwann cells was notably increased at P8 (3.187 ± 111 in the tKO versus 628 ± 21 in controls; p ≤ 0.0001). (G) The total number of sorted axons in a 1:1 relationship with Schwann cells is decreased at P2 (1.128 ± 90 in the tKO versus 2.131 ± 95 in the control; p = 0.0007). (H) The total number of Schwann cells (counted as nuclei) is increased at P8 (823 ± 37 in the tKO versus 476 ± 20 in controls; p ≤ 0.0001) and at P21 (503 ± 31 in the tKO versus 337 ± 32 in controls; p ≤ 0.0152). (I) In contrast, the number of myelinating Schwann cells is decreased at P2 (22 ± 1 in the tKO versus 134 ± 8 in controls; p ≤ 0.0001) and at P8 (153 ± 25 in the tKO versus 309 ± 11 in controls; p = 0.0013). (J) The percentage of myelinated axons is decreased at P2 (18.5 ± 3.7% in the tKO versus 54.6 ± 1.1% in controls; p ≤ 0.0001), P8 (31.6 ± 2.9% in the tKO versus 54.6 ± 1.1% in controls; p ≤ 0.0001) and, although much less, at P21 (97.9 ± 0.4% in the tKO versus 99.9 ± 0.0% in controls; p = 0.0135). For these experiments, three to four animals per genotype were used; unpaired t-test was applied for statistical analysis. (K) Markers of nonmyelin-forming Schwann cells are upregulated whereas those of myelin-forming Schwan cells are downregulated in the tKO. P2, sciatic nerves were removed and total RNA extracted. RT-qPCR with mouse-specific primers for the indicated genes was performed and normalized to 18S rRNA. Graph shows the percentage of mRNA for each gene in the tKO normalized to the control littermates. A scatter plot is shown with the results obtained, which include also the mean ± SE. (L) The same for P8. (M) The same for P21. For these experiments, four to five mice per genotype and age were used. Data were analyzed with the unpaired t-test. (N) A representative WB of protein extracts from tKO, control, and wild-type P8 nerves is shown. In the quantification, JUN protein increased in the tKO (2.88 ± 0.19 in the tKO versus 1.12 ± 0.071 in the control nerve; p = 0.004). Mpz protein was found decreased (0.55 ± 0.03 in the tKO versus 1.21 ± 0.09 in the control nerve; p = 0.0115) as was Mbp (0.62 ± 0.045 in the tKO versus 1.31 ± 0.100 in the control nerve; p = 0.012). We could not find changes in KROX-20. Densitometric analysis was done for seven to nine WB from the same number of mice and normalized to the WT. Data were analyzed with the one-way analysis of variance (ANOVA) Tukey’s test (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
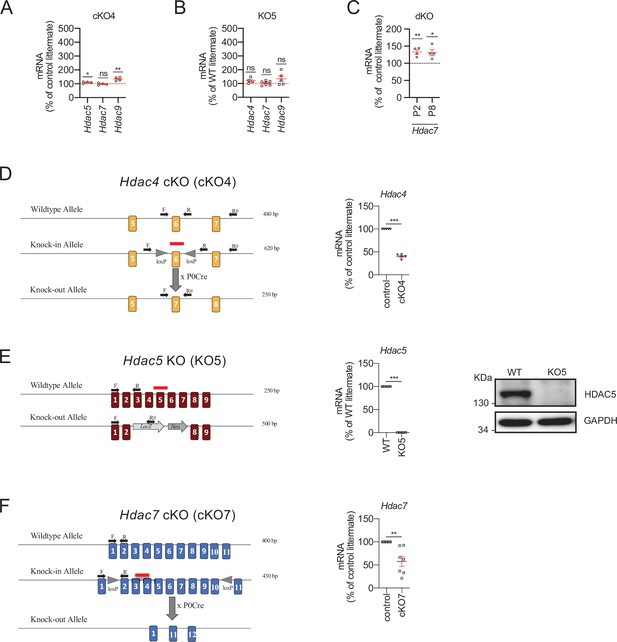
Class IIa HDAC gene expression and removal from Schwann cells.
(A) Only minor or no changes were found for in the expression of Hdac5, Hdac7, or Hdac9 in the nerves of the cKO4. (B) No changes in the expression of Hdac4, Hdac5, and Hdac9 were found in the nerves of the KO5. (C) Hdac7 compensatory overexpression is already detected in the nerves of the dKO during postnatal development (P2 and P8). RT-qPCR with mouse-specific primers for the indicated genes was performed and normalized to 18S rRNA. The scatter plot, which includes also the mean ± standard error (SE), shows the fold change in mRNA normalized to control littermates. Four to eight mice per genotype were used. Data were analyzed with the unpaired t-test. (D) To generate a Schwann cell-specific cKO4 mice, the Mpz-Cre+/− ± was crossed with a knock-in mouse with the exon 6 of Hdac4 floxed. RT-qPCR with specific primers for exon 6 (F, R) demonstrates a decrease by more than 50% in the amount of mRNA in the sciatic nerve. The absence of a complete elimination is probably due to the contribution of mRNA from other cell types. (E) For HDAC5 we used a complete HDAC5 KO mouse that has no apparent PNS phenotype. In the sciatic nerves of these mice HDAC5 was undetectable both with RT-qPCR and WB. (F) To generate a Schwann cell-specific cKO7 mice, the Mpz-Cre+/− ± was crossed with a knock-in mouse with the exons 2–10 of Hdac7 floxed. RT-qPCR with specific primers (F, R) demonstrates a decrease of about 50% in the amount of mRNA for this HDAC in the sciatic nerve. As in the case of Hdac4, the absence of a complete elimination is probably due to the contribution of mRNA from the axons and other cell types. Primer sequences are described online (Key Resources Table) (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
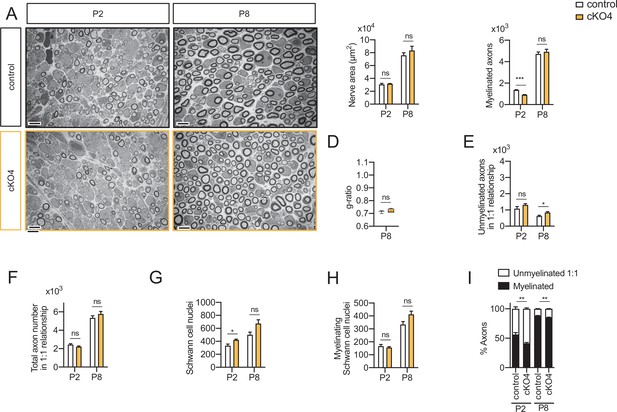
Myelin development in the cKO4 mice sciatic nerves.
Representative transmission TEM images of P2 and P8 sciatic nerves of cKO4 mice (Mpz-Cre+/−;Hdac4flx/flx) and the control (Mpz-Cre−/−;Hdac4flx/flx) littermates. Scale bar: 5 μm. (B) No statistically significant differences were observed between the area of the cKO4 nerves and control littermates. (C) The number of myelinated axons was slightly decreased at P2 (912 ± 25 in th cKO4 versus 1363 ± 29 in the control; p < 0.0001) but not at P8. (D) g ratio was not changed at P8. (E) The number of unmyelinated axons in a 1:1 relationship with Schwann cells was slightly increased at P8 (845 ± 57 in the cKO4 versus 625 ± 45 in controls; p = 0.023). (F) The total number of sorted axons in a 1:1 relationship with Schwann cells was not changed. (G) The total number of Schwann cells (counted as nucleus) is slightly increased at P2 (421 ± 12 in the cKO4 versus 333 ± 25 in controls; p = 0.013). (H) The number of myelinating Schwann cells was not changed. (I) The percentage of myelinated axons is slightly decreased at P2 (41.17 ± 1.68% in the cKO4 versus 56.29 ± 3.39% in controls; p = 0.0037) and P8 (85.37 ± 0.4% in the cKO4 versus 88.35 ± 0.41% in controls; p = 0.002). For these experiments, four to five animals per genotype were used; unpaired t-test was used for statistical analysis (*p < 0,05; **p < 0,01; ***p < 0,001; ns: no significant). See source data file one online (graphs source data) for more details. Primer sequences and antibodies are listed online (Key Resources Table).
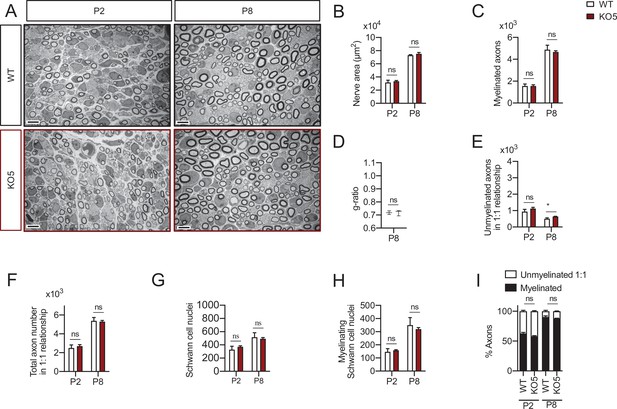
Myelin development in the KO5 mice sciatic nerves.
Representative transmission TEM images of P2 and P8 sciatic nerves of KO5 mice (HDAC5−/−) and the control (HDAC5+/+) littermates. Scale bar: 5 μm. (B) No statistically significant differences were observed between the area of the cKO4 nerves and control littermates. (C) The number of myelinated axons was not changed at P2 neither at P8. (D) g ratio was not changed at P2 neither at P8. (E) The number of unmyelinated axons in a 1:1 relationship with Schwann cells was slightly increased at P8 (632 ± 19 in the KO5 versus 503 ± 57 in controls; p = 0.044). (F) The total number of sorted axons in a 1:1 relationship with Schwann cells was not changed. (G) The total number of Schwann cells (counted as nucleus) was not changed. (H) The number of myelinating Schwann cells was not changed. (I) The percentage of myelinated axons was not changed. For these experiments three animals per genotype were used; Unpaired t-test was used for statistical analysis (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.

Myelin development in the cKO7 mice sciatic nerves.
Representative transmission TEM images of P2 and P8 sciatic nerves of cKO7 mice (Mpz-Cre+/−;Hdac7flx/flx) and the control littermates (Mpz-Cre−/−;Hdac7flx/flx). Scale bar: 5 μm. No statistically significant differences were observed between the area of the cKO4 nerves and control littermates (B), number of myelinated axons (C), g ratio (D), number of unmyelinated axons in a 1:1 relationship with Schwann cells (E), the total number of sorted axons in a 1:1 relationship with Schwann cells (F), the total number of Schwann cells (counted as nucleus) (G), and number of myelinating Schwann cells (H). The percentage of myelinated axons was not changed (I). For these experiments three to five animals per genotype were used; unpaired t-test was used for statistical analysis (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.

Myelin development in the dKO mice sciatic nerves.
Representative transmission TEM images of P2 and P8 sciatic nerves of dKO mice (Mpz-Cre+/−;Hdac4flx/flx;Hdac5−/−) and the control littermates (Mpz-Cre−/−;Hdac4flx/flx;Hdac5−/−). Scale bar: 5 μm. (B) No statistically significant differences were observed between the area of the cKO4 nerves and control littermates. (C) The number of myelinated axons is decreased at P2 (743 ± 115 in the dKO versus 1645 ± 269 in the control; p = 0.0012) but not at P8. (D) g ratio was not changed at P8. (E) The number of unmyelinated axons in a 1:1 relationship with Schwann cells was slightly increased at P8 (721 ± 32 in the dKO versus 550 ± 15 in controls; p = 0.003). (F) The total number of sorted axons in a 1:1 relationship with Schwann cells was not changed. (G) The total number of Schwann cells (counted as nucleus) was slightly increased at P8 (543 ± 18 in the dKO versus 461 ± 29 in controls; p = 0.038). (H) The number of myelinating Schwann cells was not changed. (I) The percentage of myelinated axons is decreased at P2 (34.89 ± 1.75% in the dKO versus 59.29 ± 1.97% in controls; p < 0.0001) and P8 (86.27 ± 0.62% in the dKO versus 89.47 ± 0.49% in controls; p = 0.0061). For these experiments, four to five animals per genotype were used; unpaired t-test was used for statistical analysis (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
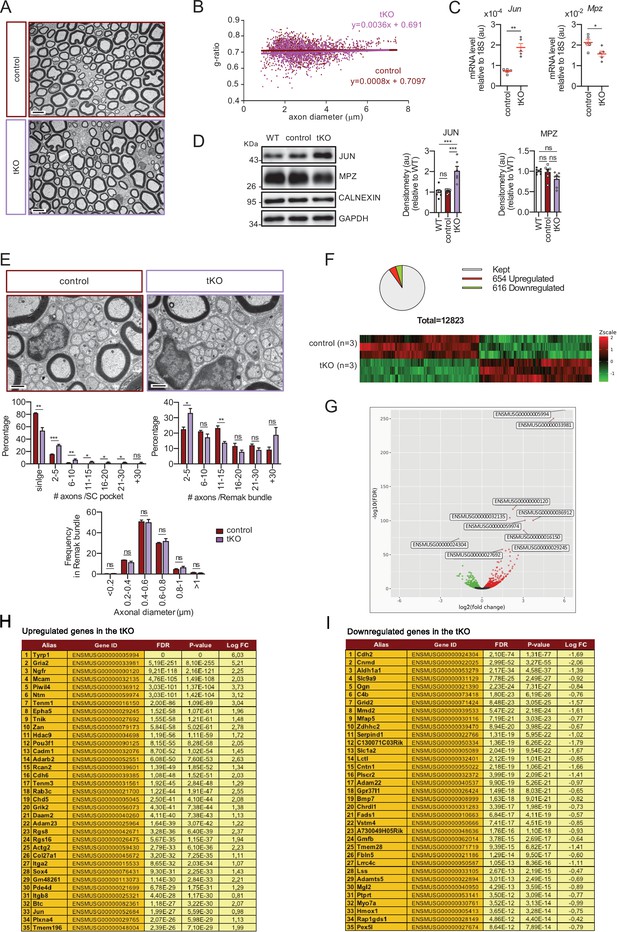
Characterization of the tKO.
(A) Representative transmission TEM image of the sciatic nerve of and adult (P60) tKO mouse and a control littermate. Scale bar: 5 μm. (B) Scatter plot of g ratio versus axon diameter. 1100 axons of 4 different mice per genotype were used. No changes in g ratio were detected. (C) mRNA for Jun remains increased in the tKO by 2.6-fold (1.88 ± 0.19 × 10−4 au in the tKO versus 0.72 ± 0.05 × 10−4 au in controls; p = 0.003) whereas Mpz was slightly decreased (1.57 ± 0.13 × 10−2 au in the tKO versus 2.13 ± 0.05 × 10−2 au in controls; p = 0.027). RT-qPCR with mouse-specific primers for the indicated genes was performed. Graph shows a scatter plot for the ΔCt (which include also the mean ± standard error [SE]) of the gene normalized to the housekeeping 18S. Five mice per genotype and age were used. Data were analyzed with the unpaired t-test. (D) JUN and MPZ protein levels. A representative Western blot of protein extracts from wild-type (C57BL/6), control and tKO sciatic nerves is shown. The densitometric analysis of six to seven different experiments normalized to WT is also shown. Data were analyzed with the unpaired t-test. Only for JUN was detected consistent changes (2.04 ± 0.22 in the tKO versus 1.05 ± 0.04 in controls; p = 0.0003) at the protein level (***p < 0.001). (E) Failed segregation of the axons in the Remak bundles of the tKO. A representative high power TEM image is shown. Morphometric analysis shows that axon diameter distribution is preserved in the tKO, but the number of axons per Remak bundle and the distribution of axon per pocket is changed. Five hundred axons from four animals per genotype were counted. Mixed model analysis of variance (ANOVA) with Bonferroni post hoc test was used for comparations. Scale bar: 1 μm. (F) Pie chart and DEG heatmap of the RNA-seq analysis of P60 showing the distribution of changed genes in the tKO. (G) Volcano plot shows that the most robustly changed genes were upregulated. ENSEMBL indentification numbers for the 10 most robustly changed genes are shown. (H) List of the 35 most upregulated genes in the adult (P60) KO classified by FDR. (I) List of the 35 most downregulated genes in the adult (P60) tKO classified by FDR (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
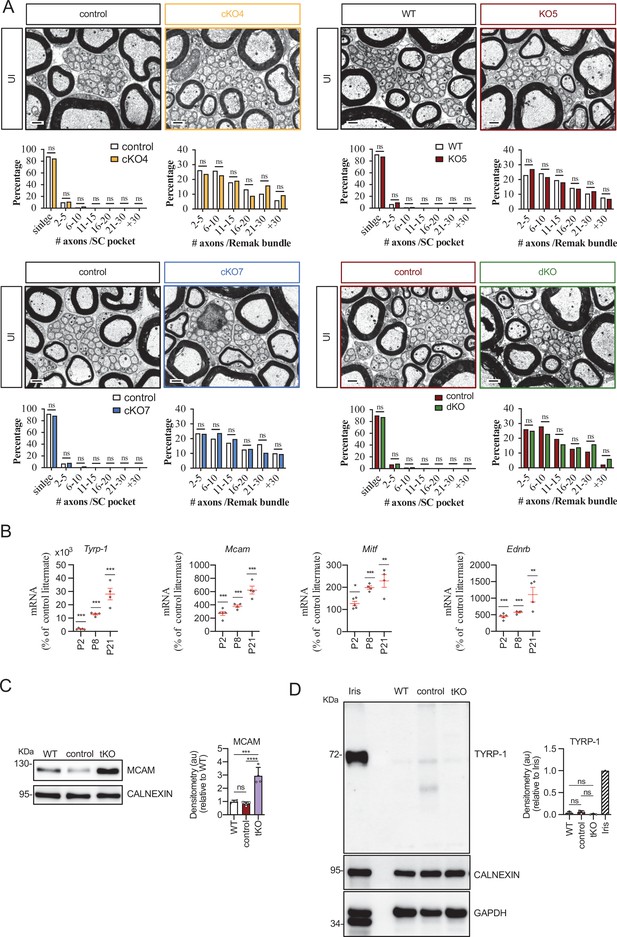
Characterizing the tKO mice.
(A) Representative TEM images and morphometric analysis of the Remak bundles of single KO and dKO show no major changes in the segregation of the small size axons in these mice. 800–1000 axons from 3 to 4 animals per genotype were counted. Mixed model analysis of variance (ANOVA) with Bonferroni post hoc test was used for comparations. Scale bar: 1 μm. (B) Melanocyte lineage genes are upregulated during early development. P2, P8, and P21 mouse sciatic nerves were removed and total RNA extracted. RT-qPCR with mouse-specific primers for the indicated genes was performed and normalized to 18S rRNA. Graph shows the percentage of mRNA for each gene in the tKO normalized to the control littermates. A scatter plot is shown with the results obtained, which include also the mean ± standard error (SE). 4/5 mice per genotype and age were used. Data were analyzed with the unpaired t-test. (C) A representative Western blot of protein extracts obtained from the sciatic nerves of P8 WT, control, and tKO mice is shown. CALNEXIN was used as a protein loading control. Densitometric analysis was done for 4 WB and normalized the WT. Data were analyzed with the one-way ANOVA Tukey’s test. (D) The mRNA for Tyrp1 is not translated to protein. A representative Western blot of protein extracts obtained from the sciatic nerves of adult WT, control, and tKO mice is shown. Iris was used as a positive control. CALNEXIN and GAPDH were used as protein loading controls. Because GAPDH is a doublet in iris, normalization was performed exclusively to CALNEXIN. Primer sequences and antibodies are listed online (Key Resources Table) (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
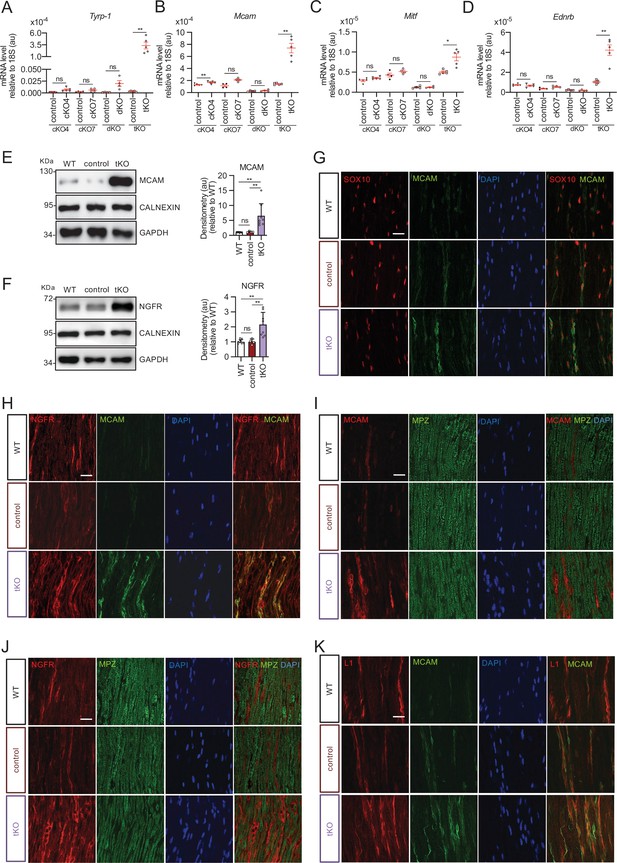
Melanocyte lineage markers are expressed by nonmyelinating Schwann cells of the Remak bundles in the sciatic nerves of the tKO.
(A) mRNA for Tyrp1 is dramatically increased by 1.081-fold in the tKO (3.35 ± 0.71 × 10−4 au in the tKO versus 0.11 ± 0.05 × 10−6 au in controls; p = 0.0092) whereas no changes were found in the cKO4, cKO7 neither dKO sciatic nerves. (B) mRNA for Mcam is also upregulated (5.13-fold) in the tKO (7.39 ± 0.79 × 10−4 au in the tKO versus 1.44 ± 0.06 × 10−4 au in controls) with only minor o no changes at all for the other genotypes. The same although less marked (1.74-fold) for Mitf (0.87 ± 0.09 × 10−5 au in the tKO versus 0.50 ± 0.02 × 10−5 au in controls; p = 0.0128) (C) and Ednrb (4.1-fold; 4.23 ± 0.52 × 10−5 au in the tKO versus 1.04 ± 0.09 × 10−5 au in controls; p = 0.0032) (D). RT-qPCR with mouse-specific primers for the indicated genes was performed. Graph shows a scatter plot for the ΔCt (which include also the mean ± standard error [SE]) of the gene normalized to the housekeeping 18S. Four to five mice per genotype were used. Data were analyzed with the unpaired t-test with Welch’s correlation. (E) MCAM protein levels in the sciatic nerves of the tKO. A representative Western blot of protein extracts from wild-type (C57BL/6), control and tKO sciatic nerves is shown. MCAM protein was increased by 7.6-fold in the tKO (9.93 ± 1.75 au in the tKO versus 1.30 ± 0.13 in controls; p = 0.0003). (F) NGFR protein was increased by 2.15-fold (2.16 ± 0.29 in the tKO versus 1.005 ± 0.09 in controls; p = 0.0003). Four to eight WB of the same number of animals per genotype were quantified. Data were analyzed with the one-way analysis of variance (ANOVA) Tukey’s test. (G) MCAM signal colocalizes with SOX10. (H) MCAM signal colocalizes with NGFR. (I) MCAM is not expressed by myelin-forming Schwann cells (MPZ+). (J) Same happens with NGFR. (K) MCAM signal colocalizes with L1cam, a marker of the nonmyelin-forming Schwann cells of the Remak bundles. P60 sciatic nerves were fixed and submitted to immunofluorescence with the indicated antibodies. Nuclei were counterstained with Hoechst. Representative confocal images of sections obtained from the sciatic nerves of wild-type (WT), control, and tKO mice are shown. Scale bar: 20 μm (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
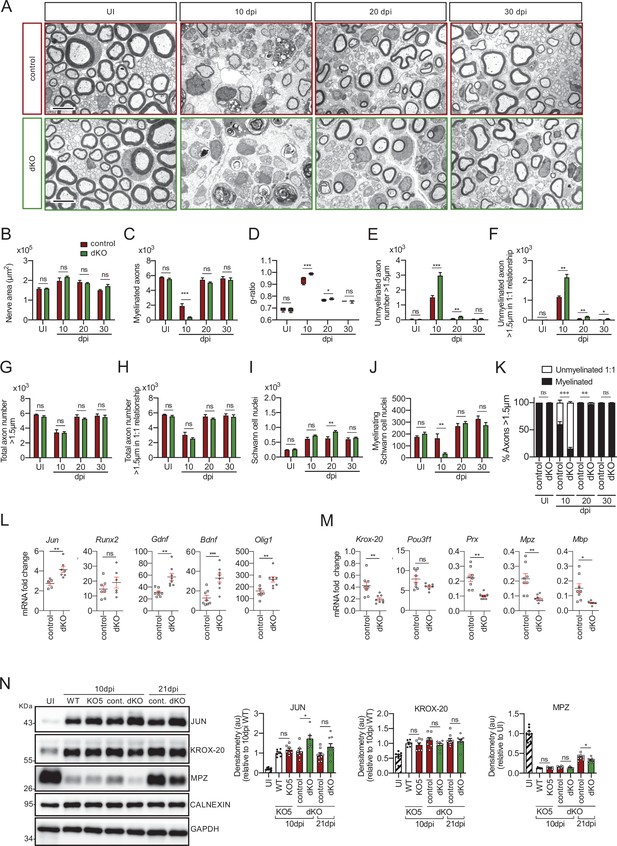
Remyelination is delayed in the nerves of the dKO mice.
(A) Representative transmission TEM images of P60 sciatic nerves uninjured (UI) and 10, 20, and 30 days post crush (dpi) of dKO (Mpz-Cre+/−; Hdac4flx/flx; Hdac5−/−) and the control (Mpz-Cre−/−; Hdac4flx/flx; Hdac5−/−) littermates are shown. Scale bar: 5 μm. (B) No statistically significant differences were observed in the area of the dKO nerves and control littermates (UI: p = 0.804; 10 dpi: p = 0.195; 20 dpi: p = 0.559; 30 dpi: p = 0.0594). (C) The number of myelinated axons is notably decreased at 10 dpi (388 ± 55 in the dKO versus 1.889 ± 330 in the control; p = 0.0005). (D) g ratio was increased at 10 dpi (0.989 ± 0.003 in the dKO versus 0.934 ± 0.015 in control [p = 0.002]) and at P21 (0.776 ± 0.003 in the dKO versus 0.767 ± 0.003 in control [p = 0.043]). (E) The number of unmyelinated axons in a 1:1 relationship with Schwann cells was notably increased at 10 dpi (2.969 ± 203 in the dKO versus 1.512 ± 119 in controls; p = 0.0007) and at 20 dpi (224 ± 25 in the dKO versus 88 ± 14 in controls; p = 0.0016). (F) The total number of unmyelinated axons in a 1:1 relationship with Schwann cells is increased at 10 dpi (2.148 ± 155 in the dKO versus 1.158 ± 56 in the control; p = 0.0011) at 20 dpi (175 ± 20 in the dKO versus 68 ± 12 in the control; p = 0.002) and at 30 dpi (63 ± 17 in the dKO versus 22 ± 5 in the control; p = 0.043). (G) No changes in the total axon number was found (UI: p = 0.157; 10 dpi: p = 0.910; 20 dpi: p = 0.349; 30 dpi: p = 0.666). (H) Neither in the total sorted axon number (UI: p = 0.193; 10 dpi: p = 0.169; 20 dpi: p = 0.294; 30 dpi: p = 0.682). (I) The total number of Schwann cells (counted as nuclei) was increased at 20 dpi (861 ± 34 in the dKO versus 630 ± 53 in controls; p = 0.0041). (J) In contrast, the number of myelinating Schwann cells was found decreased at 10 dpi (35 ± 8 in the dKO versus 164 ± 37 in controls; p = 0.0032). (K) The percentage of myelinated axons is decreased at 10 dpi (15.5 ± 2.3% in the dKO versus 60.4 ± 4.8% in controlps; p < 0.0001), 20 dpi (96.6 ± 0.4% in the dKO versus 98.8 ± 0.2% in controls; p = 0.0016) and, although much less, at P21 (98.9 ± 0.3% in the dKO versus 99.6 ± 0.1% in controls; p = 0.0482). For these experiment, three to six animals per genotype were used; unpaired t-test was applied for statistical analysis. (L) Expression of several negative regulators of myelination and repair Schwann cell markers is enhanced at 10 dpi in the sciatic nerves of the dKO: Jun (1.51-fold; p = 0.0056), Gdnf (1.85-fold; p = 0.0025), Bdnf (2.60-fold; p = 0.001), and Olig1 (1.60-fold; p = 0.008). (M) Expression of positive regulators and myelin genes is decreased at 10 dpi in the sciatic nerves of the dKO: Krox-20 (0.47-fold; p = 0.0068), Prx (0.45-fold; p = 0.001), Mpz (0.33-fold; p = 0.005), and Mbp (0.33-fold; p = 0.012). RT-qPCR with mouse-specific primers for the indicated genes was performed and normalized to 18S rRNA. The scatter plot, which include also the mean ± SE, shows the fold change of mRNA for each gene at 10 dpi normalized to the uninjured nerve. Five to eight mice per genotype were used. Data were analyzed with the unpaired t-test with Welch’s correlation. (N) A representative WB of protein extracts from dKO, control, KO5−/− and wild-type nerves is shown. In the quantification, JUN protein remains higher in the dKO at 10 dpi (1.72 ± 0.17-fold; p = 0.012) and tend to equalize at 21 dpi. MPZ protein was found decreased by 0.32 ± 0.02-fold at 21 dpi (p = 0.02), however we could not find changes in KROX-20 (KO5−/− mice were used to compare with the wild-type littermates). Densitometric analysis was done on seven to nine WB from the same number of mice and normalized to 10 dpi WT. Data were analyzed with the unpaired t-test (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
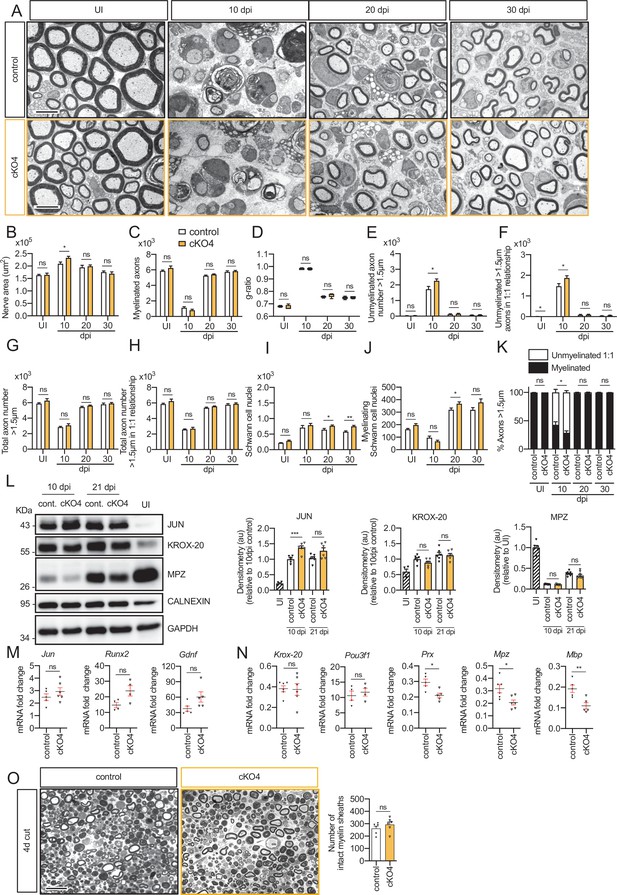
Remyelination in the cKO4 mice.
(A) Representative transmission TEM images of P60 sciatic nerves uninjured (UI) and 10, 20, and 30 days post crush (dpi) of dKO (Mpz-Cre+/−; Hdac4flx/flx) and the control (Mpz-Cre−/−;Hdac4flx/flx) littermates are shown. Scale bar: 5 μm. (B) Only slight significant statistically differences were observed in the area of the cKO4 at 10 dpi (p = 0.0372). (C) The number of myelinated axons was not changed at any point. (D) The same for g ratio. (E) The number of unmyelinated axons >1.5 μm was increased at 10 dpi in the cKO4 (1.730 ± 175 in the control versus 2.266 ± 116 in the cKO4; p = 0.024). (F) The total number of unmyelinated axons in a 1:1 relationship with Schwann cells is increased at 10 dpi (1.869 ± 115 in the cKO4 versus 1.472 ± 130 in the control; p = 0.0047). No changes in the total axon number were found (G) neither in the sorted total axon number (H). (I) The total number of Schwann cells (counted as nuclei) was increased at 20 dpi (765 ± 29 in the cKO4 versus 643 ± 41 in controls; p = 0.0041) and at 30 dpi (752 ± 34 in the cKO4 versus 575 ± 32 in controls; p = 0.0066). (J) The number of myelinating Schwann cells was slightly increased at 20 dpi (368 ± 15 in the cKO4 versus 319 ± 14 in controls; p = 0.0045). (K) The percentage of myelinated axons was decreased at 10 dpi (29.0 ± 3.2% in the cKO4 versus 42.8 ± 5.4% in controls; p = 0.0474). For these experiments, four to seven animals per genotype were used; unpaired t-test was applied for statistical analysis. (L) A representative WB of protein extracts from cKO4 and control nerves is shown. In the quantification, JUN protein was higher in the cKO4 at 10 dpi and tended to equalize at 21 dpi. No changes were found in KROX-20 or MPZ. Densitometric analysis was done on foue to six WB from the same number of mice. Data were analyzed with the unpaired t-test. To decrease the variability of standardizing for a condition with low expression, normalization was done for conditions with higher protein expression. (M) Expression of Jun, Runx2, and Gdnf at 10 dpi was not changed in the sciatic nerves of the cKO4. (N) Expression of Krox-20 and Pou3f1 was not changed, whereas Prx, Mpz, and Mbp expression was decreased in the cKO4 sciatic nerve at 10 dpi. RT-qPCR with mouse-specific primers for the indicated genes was performed and normalized to 18S rRNA. The scatter plot, which include also the mean ± standard error (SE), shows the fold change of mRNA for each gene at 10 dpi normalized to the uninjured nerve. Four to six mice per genotype were used. Data were analyzed with the unpaired t-test. (O) A representative toluidine blue staining image of 4 days cut sciatic nerve of cKO4 and control mice is shown. The quantification of intact myelin sheaths showed no changes in the cKO4. Six animals were used for the experiment. Scale bar: 10 μm. Data were analyzed with the unpaired t-test. Primer sequences and antibodies are listed online (Key Resources Table) (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.

Remyelination in the KO5 mice.
(A) Representative transmission TEM images of P60 sciatic nerves uninjured (UI) and 10, 20, 30, and 60 days post crush (dpi) of KO5 (HDAC5−/−) and the WT control littermates are shown. Scale bar: 5 μm. (B) A slight increase in the area of the KO5 at 60 dpi (p = 0.0419) was found. (C) The number of myelinated axons was not changed at any point. (D) The g ratio was found slightly decreased in at 20 dpi in the KO5 (0.75 ± 0.004 in the KO5 versus 0.78 ± 0.007 in the control; p = 0.0154). (E) The number of unmyelinated axons >1.5 μm was slightly increased at 60 dpi in the KO5 (52 ± 10 in the control versus 22 ± 6 in the KO5; p = 0.0352). The total number of unmyelinated axons in a 1:1 relationship with Schwann cells (F), the total axon number (G), the total number of sorted axons (H), the total number of Schwann cells (counted as nuclei) (I), the number of myelinating Schwann cells (J) neither the percentage of myelinated axons (K) were found changed at any point. For these experiments, three to five animals per genotype were used. Unpaired t-test was applied for statistical analysis. (L) Expression of Jun, Runx2, and Gdnf at 10 dpi was not changed in the sciatic nerves of the KO5. (M) Expression of Krox-20, Pou3f1, Prx, and Mpz was not changed, whereas Mbp expression was slightly decreased in the KO5 sciatic nerve at 10 dpi. RT-qPCR with mouse-specific primers for the indicated genes was performed and normalized to 18S rRNA. The scatter plot, which include also the mean ± SE, shows the fold change of mRNA for each gene at 10 dpi normalized to the uninjured nerve. Four to six mice per genotype were used. Data were analyzed with the unpaired t-test. (N) A representative toluidine blue staining image of 4 days cut sciatic nerve of KO5 and control mice is shown. The quantification of intact myelin sheaths showed no changes in the KO5. Scale bar: 10 μm. Six animals were used for the experiment. Data were analyzed with the unpaired t-test. Primer sequences are listed in source data section online (Key Resources Table) (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.

Myelin clearance and repair phenotype activation in the dKO mice.
(A) A representative toluidine blue staining image of 4 days cut sciatic nerve of dKO and control mice is shown. The quantification of intact myelin sheaths showed no changes. Scale bar: 10 μm. Four to five animals were used for the experiment. Data were analyzed with the unpaired t-test. (B) WB against JUN and MPZ supports that myelin clearance is normal in the dKO nerves. CALNEXIN and GAPDH were used as housekeeping. Three mice per genotype were analyzed independently by densitometry. To decrease the variability of standardizing for a condition with low expression, normalization was done for conditions with higher protein expression. Data were analyzed with the one-way analysis of variance (ANOVA) Tukey’s test. (C) Repair phenotype activation was determined by measuring the expression of marker genes and comparing with the uninjured control nerve. As is shown only a slight increase in the expression of Jun at 1 day after cut in the dKO was found (3.71 ± 0.24 in the dKO versus 2.35 ± 0.22 in the control; p = 0.0102). RT-qPCR with mouse-specific primers for the indicated genes was performed and normalized to 18S rRNA. Graph shows the percentage of mRNA for each gene in the injured nerve normalized to the uninjured controls. A scatter plot is shown with the results obtained, which include also the mean ± standard error (SE). Three to four mice per genotype were used. Data were analyzed with the unpaired t-test. Primer sequences and antibodies are listed online (Key Resources Table) (*p < 0.05; **p < 0.01; ***p < 0.001; ns: not significant). See source data file one online (graphs source data) for more details.
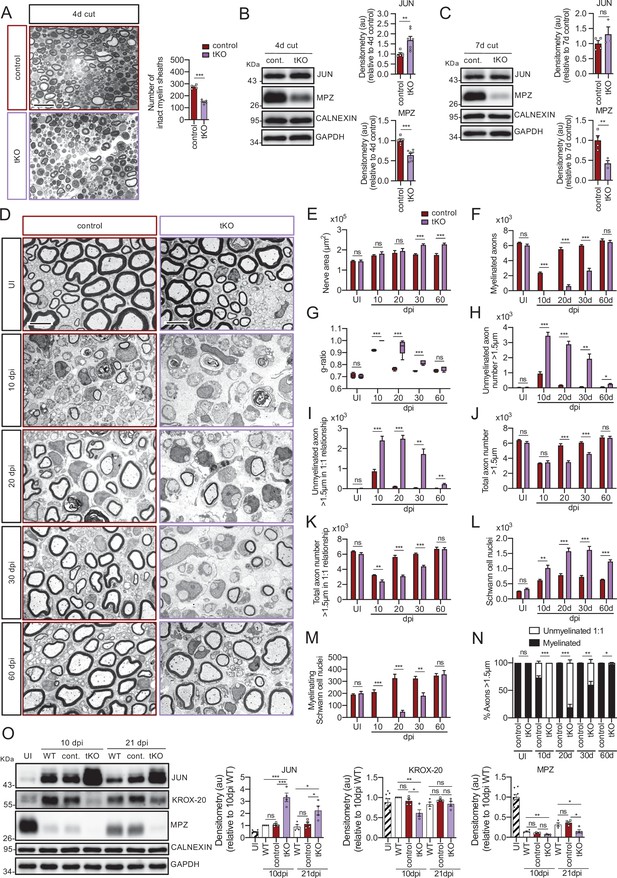
Remyelination is dramatically delayed in the tKO.
(A) Myelin clearance is accelerated in the sciatic nerves of the tKO. A representative toluidine blue staining image of 4 days cut sciatic nerve of tKO and control mice is shown. The quantification of intact myelin sheaths shows a 0.55-fold change (p < 0.0001) in the tKO (150 ± 7 intact myelin sheaths in the dKO versus 274 ± 8 in controls; p < 0.0001). Seven to eight animals per genotype were used for the experiment. Data were analyzed with the unpaired t-test. Scale bar: 10 μm. (B) WB supports an accelerated myelin clearance in the tKO. As is shown, MPZ protein is decreased (0.7-fold; p = 0.0062) in the tKO. As expected, JUN was increased. Four mice per genotype were used. (C) At 7 days post-cut the decrease in the MPZ protein was more marked (0.43-fold; p = 0.012). Three to four mice per genotype were used. CALNEXIN and GAPDH were used as housekeepings. Densitometric analysis was preformed and normalized to the controls. Data were analyzed with the unpaired t-test. (D) Representative transmission TEM images of P60 sciatic nerves uninjured (UI) and 10, 20, 30, and 60 days post crush (dpi) of tKO and the control littermates are shown. Scale bar: 5 μm. (E) No statistically significant differences were observed for the area of the tKO nerves and control littermates in the UI (p = 0.897), at 10 dpi (p = 0.456) neither at 20 dpi (p = 0.602). The nerve area of the tKO was increased at 30 dpi (1.26-fold; p = 0.0002) and at 60 dpi (1.30-fold; p = 0.0008). (F) No myelinated axons in the tKO were found at 10 dpi whereas the control had 2.383 ± 112 axons myelin (p < 0.0001). Myelinated axons were decreased at 20 dpi (606 ± 200 in the tKO versus 5.525 ± 222 in the control; p < 0.0001), at 30 dpi (2.659 ± 323 in the tKO versus 6.003 ± 125 in the control; p < 0.0001), to catch up at 60 dpi (6.458 ± 240 in the tKO versus 6.689 ± 212 in the control: p = 0.491). (G) g ratio was increased in the tKO at 10 dpi (1 ± 0 in the tKO versus 0.92 ± 0.003 in control; p < 0.0001) at 20 dpi (0.94 ± 0.027 in the tKO versus 0.77 ± 0.006 in control; p = 0.0023), and at 30 dpi (0.82 ± 0.008 in the tKO versus 0.75 ± 0.002 in control; p = 0.0009). (H) The number of unmyelinated axons >1.5 was increased at 10 dpi (3.477 ± 236 in the tKO versus 950 ± 116 in controls; p < 0.0001), at 20 dpi (2.885 ± 209 in the tKO versus 184 ± 15 in controls; p = 0.0002), at 30 dpi (1.925 ± 319 in the tKO versus 76 ± 16 in controls; p = 0.0044), and at 60 dpi (257 ± 43 in the tKO versus 69 ± 11 in controls; p = 0.010). (I) The number of unmyelinated axons >1.5 in a 1:1 relationship with Schwann was notably increased at 10 dpi (2.405 ± 209 in the tKO versus 864 ± 102 in controls; p = 0.0006), at 20 dpi (2.487 ± 170 in the tKO versus 110 ± 13 in controls; p = 0.0001), at 30 dpi (1.728 ± 250 in the tKO versus 43 ± 8 in controls; p = 0.0025), and at 60 dpi (224 ± 43 in the tKO versus 28 ± 5 in controls; p = 0.010). (J) In contrast, the total number of axons >1.5 μm was decreased at 20 dpi (3.492 ± 184 in the tKO versus 5.790 ± 228 in the control; p < 0.0001) and at 30 dpi (4.584 ± 184 in the tKO versus 6.080 ± 131 in the control; p = 0.0002) to finally catch up at 60 dpi (6.716 ± 198 in the tKO versus 6.758 ± 221 in the control; p = 0.889). (K) The total number of axons >1.5 μm sorted (in a 1:1 relationship with Schwann cells) was decreased at 10 dpi (2.405 ± 209 in the tKO versus 3.247 ± 60 in the control; p < 0.0082), at 20 dpi (3.093 ± 147 in the tKO versus 5.653 ± 233 in the control; p < 0.0001), and at 30 dpi (4.387 ± 158 in the tKO versus 6.046 ± 127 in the control; p < 0.0001). (L) The number of Schwann cells (counted as nuclei) was increased at 10 dpi (1.017 ± 95 in the tKO versus 609 ± 33 in the control; p < 0.0001), at 20 dpi (1.576 ± 100 in the tKO versus 779 ± 54 in the control; p < 0.0001), and at 30 dpi (1.618 ± 116 in the tKO versus 723 ± 47 in the control; p < 0.0001). (M) The number of myelinating Schwann cells was also decreased at 10 dpi (0 ± 0 in the tKO versus 212 ± 18 in the control; p < 0.0001), at 20 dpi (48 ± 12 in the tKO versus 326 ± 33 in the control; p < 0.0001), and at 30 dpi (181 ± 24 in the tKO versus 325 ± 17 in the control; p = 0.0011). (N) The percentage of myelinated axons is decreased at 10 dpi (0 ± 0% in the tKO versus 73.1 ± 3.1% in controls; p < 0.0001), 20 dpi (19.2 ± 5.5% in the tKO versus 98.1 ± 0.2% in controls; p < 0.0001), 30 dpi (60.3 ± 6.1% in the tKO versus 99.3 ± 0.1% in controls; p = 0.0031), and at 60 dpi (96.6 ± 0.7% in the tKO versus 99.6 ± 0.1% in controls; p < 0.0001). For these experiments, four to five animals per genotype were used; unpaired t-test was applied for statistical analysis. (O) A representative WB of protein extracts from tKO, control and wild-type nerves is shown. In the quantification, JUN protein remains higher in the tKO at 10 dpi (3.24 ± 0.35-fold; p < 0.0001) and at 21 dpi (2.25 ± 0.38-fold; p = 0.031). KROX-20 was found decreased at 10 dpi (0.61 ± 0.08-fold; p = 0.011). MPZ protein was found decreased at 21 dpi by 0.15 ± 0.04-fold dpi (p = 0.0091). Densitometric analysis was done for seven to nine WB from the same number of mice and normalized to 10 dpi WT. Data were analyzed with the unpaired t-test (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
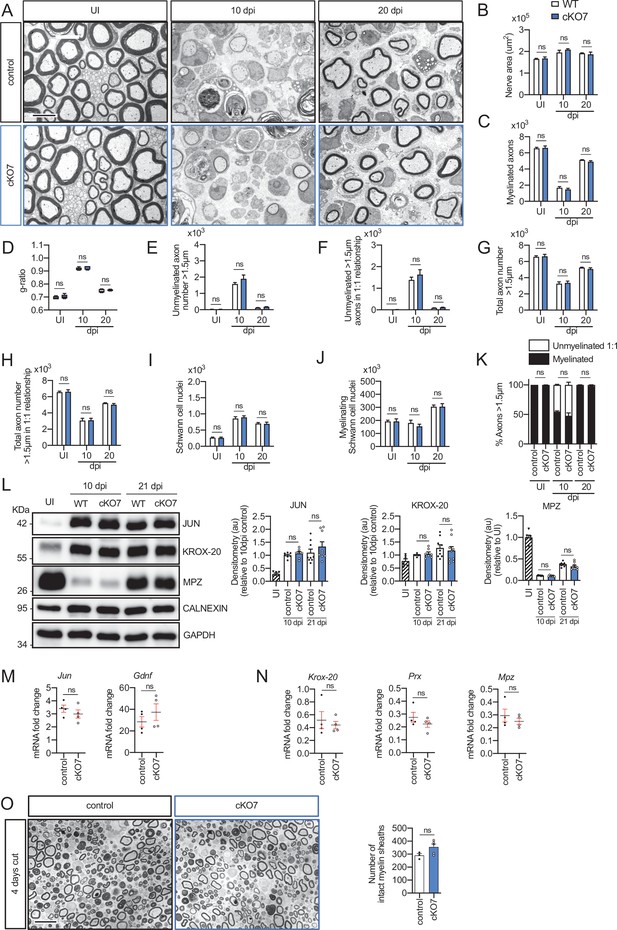
Remyelination in the cKO7 mice.
(A) Representative transmission TEM images of P60 sciatic nerves uninjured (UI) and 10 and 20 days post crush (dpi) of cKO7 (Mpz-Cre+/−;Hdac7 flx/flx) and the WT control littermates (Mpz-Cre−/−;Hdac7 flx/flx) are shown. Scale bar: 5 μm. No changes were found at any point in the nerve area (B), the number of myelinated axons (C), g ratios (D), number of unmyelinated axons >1.5 μm (E), the total number of unmyelinated axons >1.5 μm in a 1:1 relationship with Schwann cells (F), the total axon number (>1.5 μm) (G), the total number of sorted axons (H), the total number of Schwann cells (counted as nuclei) (I), the number of myelinating Schwann cells (J) neither the percentage of myelinated axons (K). For these experiments, three to five animals per genotype were used. Unpaired t-test was applied for statistical analysis. (L) A representative Western blot of protein extracts obtained from sciatic nerves UI, 10 and 21 days post crush (pdi) is shown. Densitometric quantification shows no differences between phenotypes. Six to nine mice were used for quantification. To decrease the variability of standardizing for a condition with low expression, normalization was done for conditions with higher protein expression. (M) No changes were found in the mRNA for Jun and Gdnf at 10 dpi. (N) No changes were found for Krox-20, Prx, and Mpz at 10 dpi. RT-qPCR with mouse-specific primers for the indicated genes was performed and normalized to 18S rRNA. Graph shows the percentage of mRNA for each gene in the crush injured (10 dpi) nerve normalized to the uninjured controls. A scatter plot is shown with the results obtained, which include also the mean ± standard error (SE). Four mice per genotype were used. Data were analyzed with the unpaired t-test. (O) A representative toluidine blue staining image of 4 days-cut sciatic nerve of cKO7 and control mice is shown. The quantification of intact myelin sheaths shows no changes in the cKO7. Scale bar: 10 μm. Three to five animals were used for the experiment. Data were analyzed with the unpaired t-test. Primer sequences and antibodies are listed online (Key Resources Table) (*p < 0.05, **p < 0.01, ***p < 0.001; ns: not significant). See source data file one online (graphs source data) for more details.
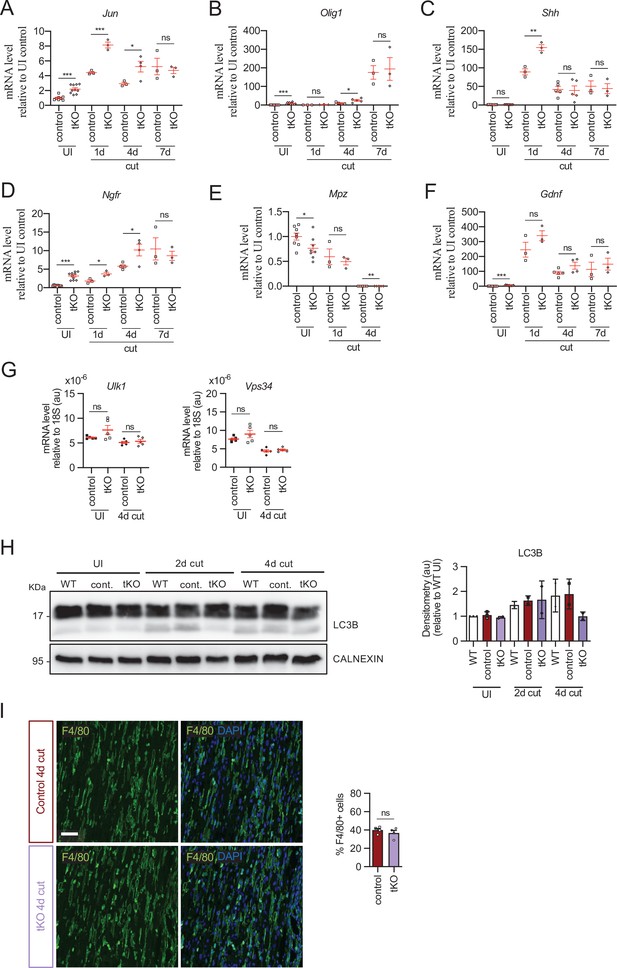
Repair Schwann cell phenotype and myelin removal in the injured tKO.
tKO Schwann cells express efficiently markers of the repair phenotype after a cut experiment. In some time points, Jun (A), Olig1 (B), Shh (C), and Ngfr (D) were even more expressed in the tKO than in the control nerves. Mpz was decreased in the UI and at 4 days post cut (E). No changes were found in Gdnf (F). (G) Autophagy gene expression at 4 days after cut was not modified in the tKO nerves. A cut experiment was performed (P60) and sciatic nerves removed at 1, 4, and 7 days postinjury. RT-qPCR with mouse-specific primers for the indicated genes was performed and normalized to 18S rRNA. Graph shows the percentage of mRNA for each gene in the cut injured (4-day cut) nerve normalized to the uninjured controls. A scatter plot is shown with the results obtained, which include also the mean ± standard error (SE). Three to five animals per time point and genotype were used, except for uninjured (10–11 nerves). (H) We also found no changes in the activation of LC3B in the tKO nerves by WB during myelin clearance. Data were analyzed with the one-way analysis of variance (ANOVA) Tukey’s test. (I) No changes were found in the number of macrophages in the tKO nerves. Representative confocal images of sections obtained from the sciatic nerves of control and tKO mice 4 days after cut are shown. Macrophages were stained with anti F4/80 antibody. Scale bar: 20 μm. For the quantification, four animals per genotype were used. Antibodies and primers used are listed in source data section online (Key Resources Table). Data were analyzed with the unpaired t-test (*p < 0.05, **p < 0.01, ***p < 0.001; ns: not significant). See source data file one online (graphs source data) for more details.
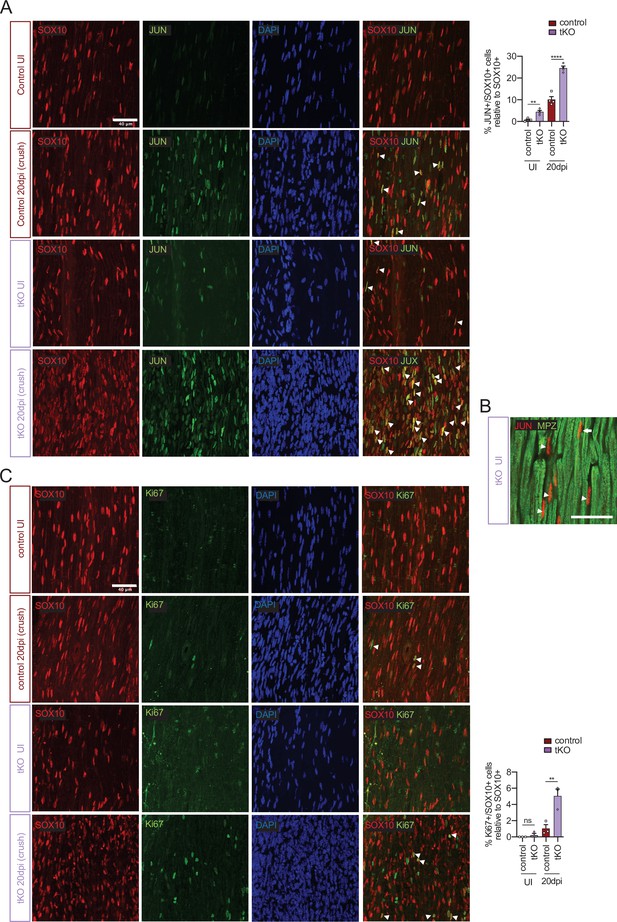
Increased Jun and Schwann cell proliferation in the tKO sciatic nerve.
(A) tKO mice sciatic nerves showed increased numbers of Schwann cells (SOX10+) expressing JUN both before and 20 days post crush. (B) Most of the JUN-positive cells not colocalize with MPZ (arrowheads), suggesting they are Remak Schwann cells. However, some of them colocalize with MPZ, showing JUN is also expressed by subgroup of myelinating Schwann cells (arrow). (C) tKO mice sciatic nerves show increased numbers of Schwann cell (SOX10+) proliferating (Ki67+) at 20 dpi. Uninjured and 20 dpi crushed sciatic nerves (P60) were fixed and submitted to immunofluorescence with the indicated antibodies. Nuclei were counterstained with Hoechst. Representative confocal images of sections obtained from the sciatic nerves of wild-type (WT), control and tKO mice are shown. Scale bar: 40 μm. For the quantification, four animals per genotype were used. Data were analyzed with the one-way analysis of variance (ANOVA) Tukey’s test. Antibodies used are listed online (Key Resources Table) (*p < 0.05, **p < 0.01 ***p < 0.001; ns: not significant). See source data file one online (graphs source data) for more details.
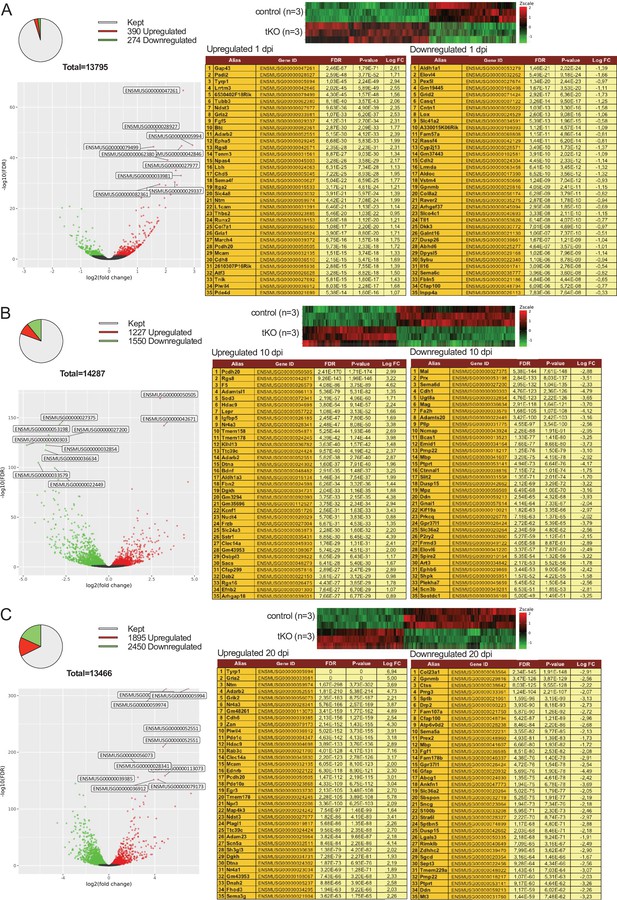
Evolution of sciatic nerve gene expression profile at 1, 10, and 20 days post crush (dpi).
The Pie chart, DEG heatmap, volcano plot and list of the 35 most upregulated and downregulated genes in the tKO classified by FDR at 1 dpi (A), 10 dpi (B), and 20 dpi (C) are shown. Data obtained from the RNA-seq analysis of three animals per condition. See source data file two online (RNA-seq source data) for more details.
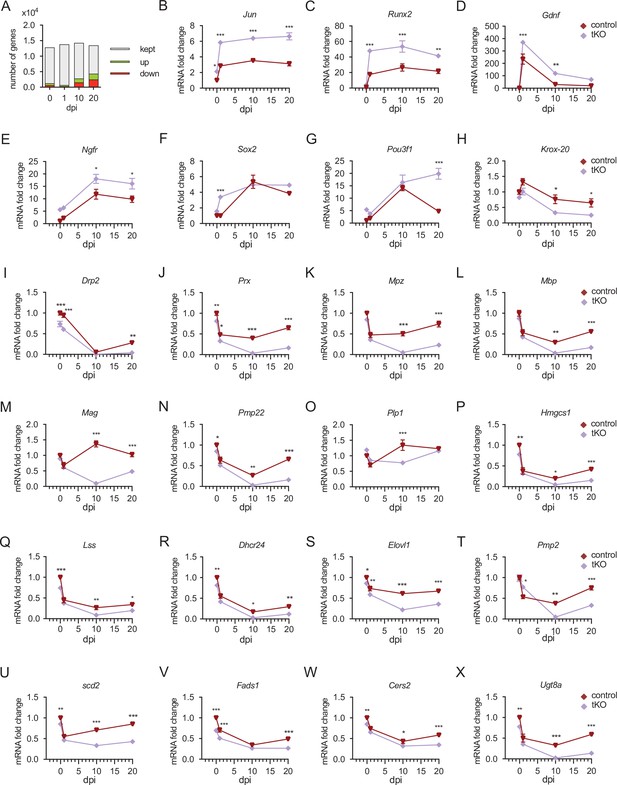
Time course analysis of myelination-relevant gene expression during nerve regeneration from RNA-seq.
(A) Bar chart showing an increase in the number of DE genes at different time points after nerve crush. In the injured tKO nerve 1.270 genes are DE being 390 upregulated and 274 downregulated. At 1 dpi, the number of genes DE genes decreased to 664, being 390 upregulated and 274 downregulated. At 10 dpi, there were 2.777 genes DE, being 1.227 upregulated and 1.550 downregulated. The bigger differences were found at 20 dpi, with up to 4.345 DE genes, being 1.895 upregulated and 2.450 downregulated. (B–F) The expression of markers of nonmyelin-forming Schwann cells, such as Jun, Runx2, Gdnf, Ngfr, and Sox2, is enhanced in the sciatic nerves of the tKO mice (G) Pou3f1 is induced in the control and tKO animals by more than 10-fold at 10 dpi. Then, at 20 dpi it drops in the control nerves but still grows up in the tKO (up to 15-fold). (H–O) Krox-20 and myelin protein genes are downregulated in the tKO nerves. (P–R) Different genes of the sterol branch of the mevalonate pathway (Hmgcs, Lss, and Dhcr24) are downregulated in the tKO. We also found downregulated genes encoding for enzymes involved in the elongation, (Elovl1) (S), transport (Pmp2) (T), and the insertion of double bonds (Scd2 and Fads1) (U, V) into fatty acids. Finally, we found downregulated Cers2 and Ugt8a, genes involved in the synthesis of sphingomyelin and galactosyl-ceramide, respectively (W, X). Level of mRNA for each gene (as FPKM) and genotype was normalized to the level of the uninjured control nerve and expressed as fold change. The sciatic nerve of three mice per genotype and timepoint were used to perform the RNA-seq (see Material and methods). Two-way analysis of variance (ANOVA) test was used for the analysis (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
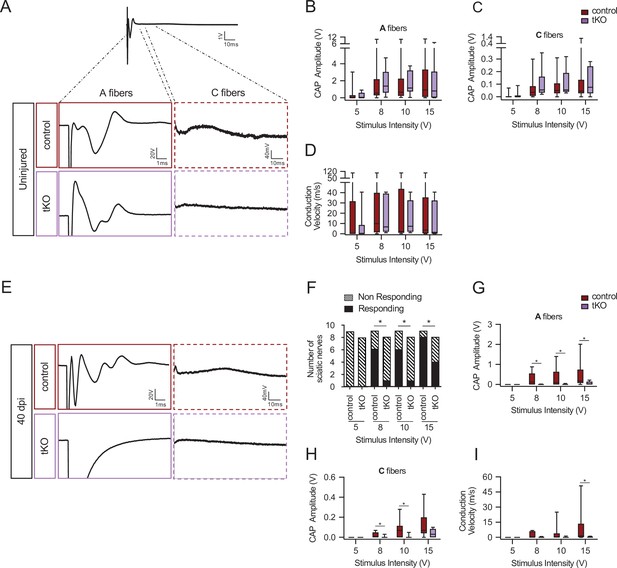
Remyelination failure in the tKO hampered nerve impulse conduction.
(A) Sample recordings of compound action potential (CAP) in uninjured sciatic nerves of control and tKO mice showing the waveform components corresponding to myelinated (A-fibers) and unmyelinated (C) fibers. (B, C) Waveform component of A-fibers (B) and C-fibers (C) showed similar amplitude in both genotypes for stimulation with pulses of increasing intensity (5, 8, 10, and 15 V). (D) Nerve conduction velocity was also preserved in the tKO mice. (E) Sample recordings of CAPs obtained in a control and a tKO sciatic nerve 40 days after nerve crushing. (F) The number of nerves that responded with a detectable CAP after the stimulation with increasing intensity. (G) Amplitude of CAP corresponding to A-fibers was significantly smaller in tKO than in control nerves for stimulation with 8, 10, and 15 V. (H) Amplitude of C-fiber component was significantly smaller in tKO than in control nerves for stimulation with 8 and 10 V. (I) Nerve conduction velocity was significantly reduced after injury in both genotypes, being significantly lower in tKO than in control nerves for stimulation at 15 V. In this set of experiments, the whole length of a sciatic nerve was exposed from its proximal projection (L4 spinal cord) to its distal branches in deeply anesthetized mice. Compound action potentials (CAPs) were evoked by electrical stimulation of increasing intensity (5, 8, 10, and 15 V, 0.03-ms pulse duration). The maximum amplitude of the A- and C-fiber components of CAP electrical signal, and their mean nerve conduction velocity were measured. Seven to 18 animals per genotype and condition were used. Mann–Whitney’s U was used for nonparametric paired comparisons and chi-squared test was used for statistical comparations (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
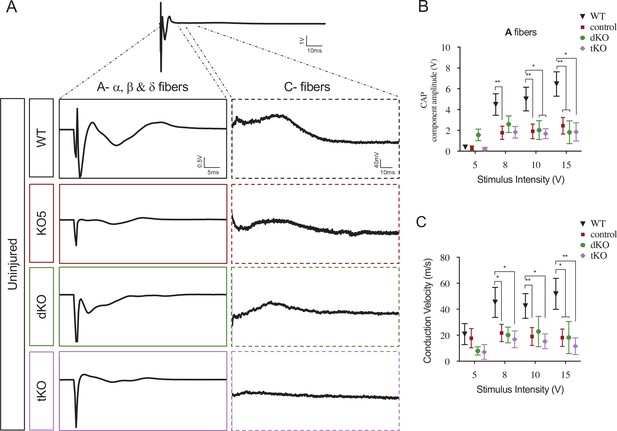
HDAC5 elimination decreases nerve conduction velocity and voltage amplitude.
(A) Sample recordings of compound action potential (CAP) in uninjured sciatic nerves of WT, KO5, dKO, and tKO mice showing the waveform components corresponding to myelinated (A-fibers) and unmyelinated (C) fibers. (B) Waveform component of A-fibers showed a decreased amplitude for all genotypes (compared to WTs) at pulses of 8, 10, and 15 V. (C) The same for nerve conduction velocity. In this set of experiments, the whole length of a sciatic nerve was exposed from its proximal projection (L4 spinal cord) to its distal branches in deeply anesthetized mice. CAPs were evoked by electrical stimulation of increasing intensity (5, 8, 10, and 15 V, 0.03-ms pulse duration). The maximum amplitude of the A-fiber components of CAP electrical signal, and their mean nerve conduction velocity were measured. Seven to 18 animals per genotype were used. Mann–Whitney’s U was used for nonparametric paired comparisons (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
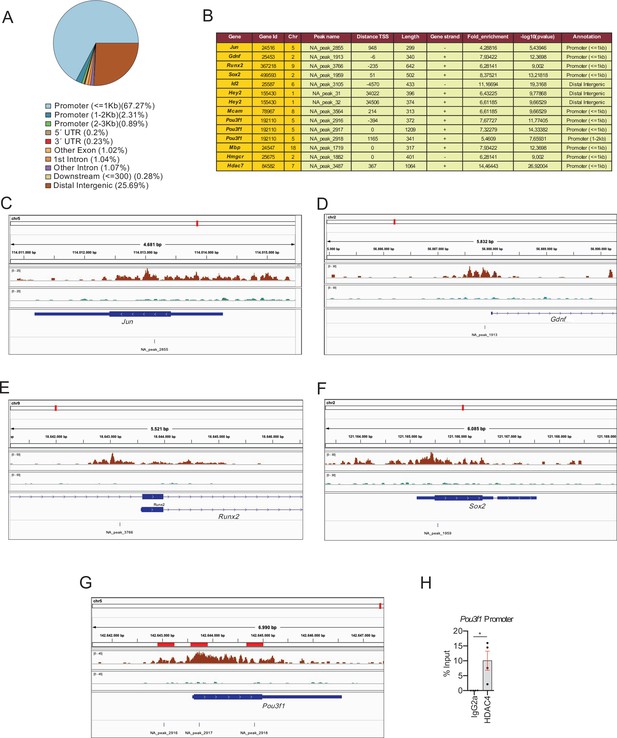
HDAC4 binds to the promoter of pivotal genes for myelin development.
Cultured rat Schwann cells were incubated with 1 mM dbcAMP to shuttle HDAC4 into the nucleus, and ChIP-Seq analysis performed on the crosslinked chromatin, with anti-HADC4 antibody. (A) 3.932 HDAC4 peaks were found in the rat genome. Most of these peaks (67.27%) were located in the promoter regions (≤1 kb from the TSS). (B) A table with the localization of some of these peaks (a complete list can be found in source data file three online [ChIP-Seq peaks source data]). (C–E) ChIP-Seq signal analysis confirmed our previous results (Gomis-Coloma et al., 2018) showing that HDAC4 binds to the promoters of Jun, Gdnf, and Runx2. (F) HDAC4 was also found bound to the promoter region of Sox2. (G) Three peaks (NA_peaks 2916, 2917, and 2918) near the TSS of Pou3f1 were also found. (H) The binding of HDAC4 to Pou3f1 gene was confirmed by ChIP-qPCR. Four different experiments from four distinct cultures were used. Data were analyzed with the Mann–Whitney test (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
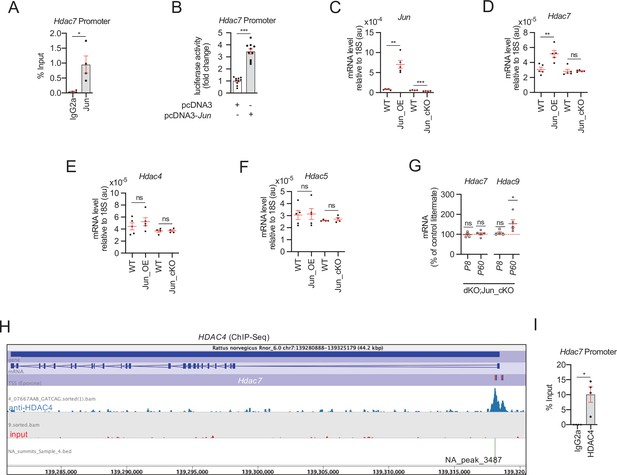
HDAC7 compensatory overexpression is induced by JUN.
(A) ChIP-qPCR of dbcAMP treated rat Schwann cells with anti-JUN antibody showed that this transcription factor is bound to the promoter of Hdac7. Four different experiments from four distinct cultures were used. Data were analyzed with the Mann–Whitney test. (B) A 1.189 fragment of the mouse Hdac7 promoter containing a conserved JUN binding consensus sequence was PCR amplified and cloned into the pGL3 luciferase reporter vector. HEK293 cells were transfected with this construct and the pcDNA3 (empty vector) or pcDNA3 Jun. As is shown JUN induced the luciferase activity by 3.4 ± 0.22-fold (p < 0.0001; n = 10). Unpaired t-test with Welch’s correlation was used for statistical comparison. (C) Levels of mRNA for Jun in the nerves of Jun_OE and Jun_cKO mice. (D) Hdac7 expression is enhanced in the sciatic nerves of the Jun_OE mice (5.16 ± 0.46 × 10−5 in the Jun_OE versus 3.09 ± 0.29 × 10−5 in the WT; p = 0.005) but does not change in the Jun_cKO mice. (E) Hdac4 expression in sciatic nerves does not change in Jun_OE and Jun_cKO mice. (F) Hdac5 expression in sciatic nerves does not change in Jun_OE and Jun_cKO mice. (G) Removal of Jun from Schwann cells in the dKO (dKO;Jun_cKO genotype) prevents Hdac7 compensatory overexpression. Interestingly, Hdac9 expression is induced in these mice (see discussion). RT-qPCR with mouse-specific primers for the indicated genes was performed. The scatter plot, which include also the mean ± standard error (SE), shows the expression of each gene normalized to the housekeeping 18S. Four to five mice per genotype were used. Data were analyzed with the unpaired t-test with Welch’s correlation. (H) A peak of HDAC4 (NA_peak 3487) was found on the Hdac7 promoter in the ChIP-Seq experiemt. (I) ChIP-qPCR confirmed that HDAC4 is bound to the promoter of Hdac7. Four different experiments from four distinct cultures were used. Data were analyzed with the Mann–Whitney test (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.

The proximal promoter region of the Hdac7 has a JUN consensus binding sequence.
(A) The alignment of the proximal promoter region for human, mouse, and rat is shown. A JUN consensus binding sequence in the promoter of the mouse Hdac7 gene was identified (yellow). The consensus sequence is conserved in rat and humans. The transcription starting site defined for rat is shown in a red box. In blue, the sequence of the primers used to amplify and clone the mouse Hdac7 promoter region are shown. In purple are the sequences of the primers used for the ChIP-qPCR with anti-JUN and anti-HDAC4. (B) Density of reads in the ChIP-Seq and mapping of the JUN consensus sequence and primer position in the Hdac7 gene.
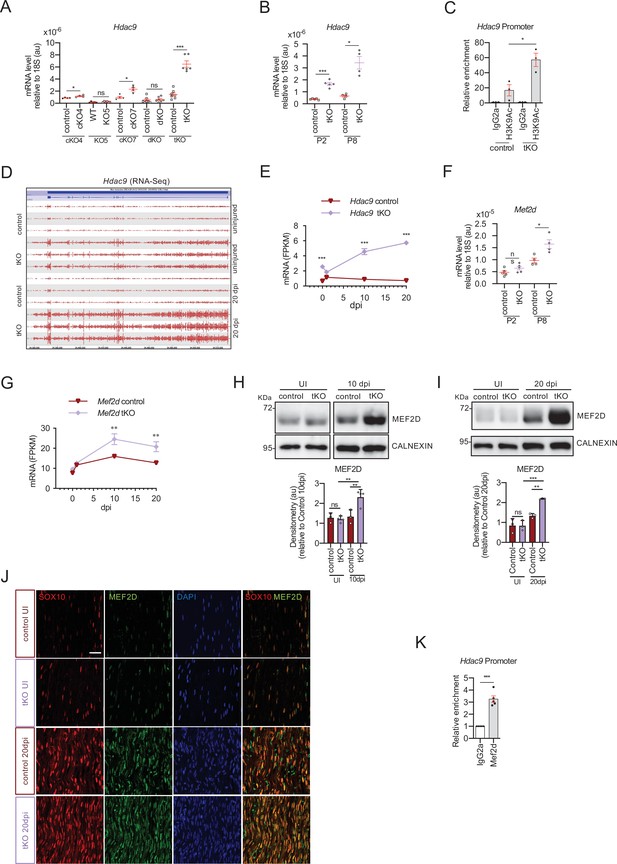
MEF2D mediates Hdac9 de novo expression in the tKO.
(A) Hdac9 expression is notably induced in the sciatic nerves of the adult (P60) tKO mice (6.48 ± 0.53 × 10−6 au the tKO versus 1.46 ± 0.28 × 10−6 in the control; p < 0.0001). Only minor changes were observed in the cKO4 and cKO7. Four to eight mice per genotype were used. Unpaired t-test was used for comparations. (B) Hdac9 expression is increased from early postnatal development of the tKO nerve. At P2 we found 1.67 ± 0.13 × 10−6 au in the tKO versus 0.39 ± 0.03 × 10−6 in the controls (p < 0.0001) and at P8 we found 3.43 ± 0.52 × 10−6 au in the tKO versus 0.65 ± 0.09 × 10−6 in the controls (p < 0.0001). RT-qPCR with mouse-specific primers for Hdac9 was performed. The scatter plot, which include also the mean ± standard error (SE), shows the expression of Hdac9 normalized to the housekeeping 18S. Four to five mice per genotype were used. Data were analyzed with the unpaired t-test with Welch’s correlation. (C) ChIP-qPCR with anti-H3K9Ac of adult (P60) sciatic nerves of tKO and control mice. Three different experiments of four to five animals per genotype are shown. Data were normalized to the IgG value as shown as relative enrichment. Unpaired t-test was used for comparations. (D) Alignment of the reads of the RNA-seq from three individual sciatic nerves of control and three tKO mice, both uninjured and at 20 days post crush (20 dpi). Hdac9 gene is transcribed at detectable levels in the sciatic nerve of the uninjured tKO mice, whereas it is almost nondetectable in the control sciatic nerves. The tKO mice (but not the controls) increase additionally the expression of Hdac9 gene during remyelination (20 dpi). (E) mRNA levels of Hdac9 (as FPKMs) at 0, 1, 10, and 20 days post crush (dpi) in the RNA-seq experiment. Two-way analysis of variance (ANOVA) was used for statistical comparation. (F) Mef2d expression is increased early in development (P8) in tKO nerve (1.65 ± 0.18 in the tKO versus 0.97 ± 0.10 in controls; p = 0.025). RT-qPCR with mouse-specific primers for Mef2d was performed. The scatter plot, which include also the mean ± standard error (SE), shows the expression normalized to the housekeeping 18S. Four to five mice per genotype were used. Data were analyzed with the unpaired t-test with Welch’s correlation. (G) mRNA levels of Mef2d (as FPKMs) at 0, 1, 10, and 20 days post crush (dpi) in the RNA-seq experiment. (H) A representative WB of protein extracts from tKO, control, and wild-type nerves at 10 dpi is shown. In the quantification, MEF2D protein was increased in the tKO nerves (2.31 ± 0.19 au in the tKO versus 1.33 ± 0.19 in controls; p < 0.0069). (I) Same for 20 dpi (2.19 ± 0.03 au in the tKO versus 1.33 ± 0.11 in controls; p < 0.0073). Densitometric analysis was done for three to four WB from the same number of mice and normalized to the control 20 dpi. Data were analyzed with the unpaired t-test. (J) MEF2D colocalizes with the transcription factor SOX10+, suggesting that it is expressed by Schwann cells. P60 sciatic nerves were fixed and submitted to immunofluorescence with the indicated antibodies. Nuclei were counterstained with Hoechst. Representative confocal images of sections obtained from the sciatic nerves of control and tKO mice are shown. Scale bar: 20 μm. (K) MEF2D binds to the Hdac9 promoter in the tKO. ChIP-qPCR of 20 dpi nerves of tKO mice was performed using an anti-MEF2D-specific antibody. Five different experiments from four to five mice per genotype were performed. Data were analyzed with the unpaired t-test (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.

Hdac9 gene expression regulation.
The in vivo overexpression of JUN in Schwann cells (Jun_OE mice) did not modify the expression of Hdac9 gene in the PNS. RT-qPCR with mouse-specific primers for Hdac9 was performed and normalized to 18S rRNA. Graph shows the relative expression of the mRNA normalized to the expression of 18S. A scatter plot is shown with the results obtained, which include also the mean ± standard error (SE). Five mice per genotype were used. Data were analyzed with the unpaired t-test. Primer sequences are listed online (Key Resources Table) (*p < 0.05; **p < 0.01; ***p < 0.001; ns: no significant). See source data file one online (graphs source data) for more details.
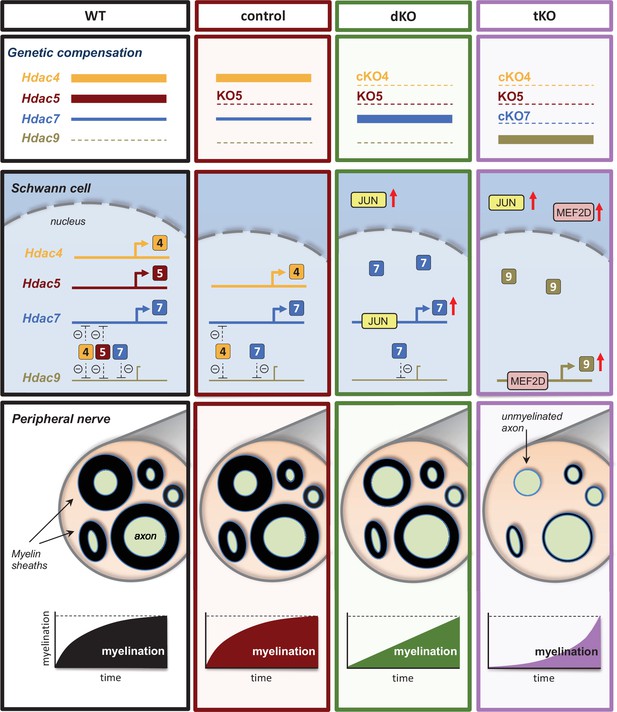
A graphical summary of the proposed model: In wild-type nerves (WT), the expression of Hdac4, Hdac5, and Hdac7 allows myelin formation and blocks the expression of Hdac9.
The removal of Hdac5 (control) has no effects on myelination neither Hdac9 expression. In the nerves of the dKO, JUN induces the compensatory overexpression of HDac7, allowing delayed myelination but having no effect on Hdac9 gene expression. The simultaneous elimination of Hdac4, Hdac5, and Hdac7 (tKO) induces the overexpression of Mef2d, which binds to the promoter and induce the compensatory expression of Hdac9, which after a long delay induces myelination.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus, male and female) | cKO4 mouse Mpz < Cre/+>;Hdac4< f/f> | Potthoff and Olson, 2007a | Hdac4tm2.1Eno | C57BL/6 background, RRID:MGI:4418117 |
| Strain, strain background (Mus musculus, male and female) | KO5 mouse Hdac5(-/-) | Chang et al., 2004 | Hdac5tm1Eno | C57BL/6 background, RRID:MGI:3056065 |
| Strain, strain background (Mus musculus, male and female) | cKO7 mouse Mpz < Cre/+>;Hdac7< f/f> | Chang et al., 2006 | Hdac7tm2Eno | C57BL/6 background, RRID:MGI:3693628 |
| Strain, strain background (Mus musculus, male and female) | Jun_OE mouse Mpz < Cre/+>;Rosa26< flox-stop-Jun/+> | Fazal et al., 2017 | Gt(ROSA)26Sortm15(Jun)Rsky | C57BL/6 background, RRID:MGI:6478892 |
| Strain, strain background (Mus musculus, male and female) | Jun_cKO mouse Mpz < Cre/+>;Jun < f/f> | Behrens et al. (2002) | Juntm4Wag | C57BL/6 background, RRID:MGI:2445420 |
| Strain, strain background (Mus musculus, male and female) | Mpz < Cre/+> | Jackson Laboratory | B6N.FVB-Tg(Mpz-cre)26Mes/J | Mpz-Cre mouse C57BL/6J background, RRID:IMSR_JAX:017927 |
| Antibody | anti- CANELXIN (rabbit polyclonal) | Enzo Life Sciences | Cat# ADI-SPA-860-D; RRID:AB_2038898 | WB (1:1000) |
| Antibody | anti- JUN (rabbit monoclonal) | Cell Signaling | Cat #9165; RRID:AB_2130165 | WB (1:1000), IF (1:800) |
| Antibody | anti- KROX-20 (rabbit polyclonal) | Millipore | Cat# ABE1374; RRID:AB_ 2715555 | WB (1:500) |
| Antibody | anti- Ki67 (rabbit polyclonal) | Abcam | Cat# ab15580; RRID:AB_443209 | IF (1:100) |
| Antibody | anti- F4/80 (rat monoclonal) | BioRad | Cat# MCA497GA; RRID:AB_323806 | IF (1:100) |
| Antibody | anti- GAPDH (rabbit polyclonal) | Sigma-Aldrich | Cat# G9545; RRID:AB_796208 | WB (1:5000) |
| Antibody | anti- HDAC4 (mouse monoclonal) | Sigma-Aldrich | Cat# H0163; RRID:AB_477042 | ChIP (10 mg) |
| Antibody | anti- HDAC5 (mouse monoclonal) | Santa Cruz | Cat# sc-133106; RRID:AB_2116793 | WB (1:1500) |
| Antibody | anti- IgG2a (mouse monoclonal) | Sigma-Aldrich | Cat# M7769; RRID:AB_1163540 | ChIP (10 mg) |
| Antibody | anti- L1 (rat monoclonal) | Chemicon International | Cat# MAB5272; RRID:AB_2133200 | IF (1:50) |
| Antibody | anti- LC3B (rabbit polyclonal) | Sigma-Aldrich | Cat# L7543; RRID:AB_796155 | WB (1:1000) |
| Antibody | anti- MCAM (rabbit monoclonal) | Origene | Cat# TA303592; RRID:AB_2143390 | WB (1:1000) IF (1:200) |
| Antibody | anti- MPZ (chicken polyclonal) | AvesLab | Cat# PZ0; RRID:AB_2313561 | WB (1:1000) IF (1:1000) |
| Antibody | anti- p75NTR (NGFR) (rabbit polyclonal) | Covance | Cat# PRB-602C; RRID:AB_291707 | WB (1:1000) |
| Antibody | anti- NGFR (mouse monoclonal) | Thermo Fisher Scientific | Cat# MA5-13314; RRID:AB_10982037 | IF (1:100) |
| Antibody | anti- SOX10 (goat polyclonal) | R&D Systems | Cat# AF2864; RRID:AB_442208 | IF (1:100) |
| Antibody | anti- TYRP1 (rabbit polyclonal) | Sigma-Aldrich | Cat# SAB2102617 RRID:AB_10611135 | WB (1:1000) |
| Antibody | anti- Rabbit IgG, HRP-linked (goat polyclonal) | Cell Signaling | Cat# 7074; RRID:AB_2099233 | WB (1:2000) |
| Antibody | anti- Mouse IgG, HRP-linked (horse polyclonal) | Cell Signaling | Cat# 7076; RRID:AB_330924 | WB (1:2000) |
| Antibody | anti- Chicken IgY (IgG) (rabbit polyclonal) | Sigma-Aldrich | Cat# A9046; RRID:AB_258432 | WB (1:2000) |
| Antibody | Cy3 anti-Rabbit IgG (H + L) (donkey polyclonal) | Jackson Immuno Research Labs | Cat# 711-165-152; RRID:AB_2307443 | IF (1:500) |
| Antibody | anti-Goat Alexa 555 Conjugated (donkey polyclonal) | Molecular Probes - Thermo Fisher | Cat# A21432; RRID:AB_2535853 | IF (1:1000) |
| Antibody | anti-Rabbit Alexa 488 Conjugated (donkey polyclonal) | Molecular Probes - Thermo Fisher | Cat# A21206; RRID:AB_2535792 | IF (1:1000) |
| Antibody | anti-Chicken Alexa 488 Conjugated (donkey polyclonal) | Jackson Immuno Research Labs | Cat# 703-545-155; RRID:AB_2340375 | IF (1:1000) |
| Antibody | anti-Rat Alexa 488 Conjugated (donkey polyclonal) | Molecular Probes - Thermo Fisher | Cat# A21208; RRID:AB_141709 | IF (1:1000) |
| Antibody | anti-Rat Alexa 555 Conjugated (goat polyclonal) | Molecular Probes - Thermo Fisher | Cat# A21434; RRID:AB_2535855 | IF (1:1000) |
| Antibody | Cy3 anti-Mouse IgG (H + L) (donkey polyclonal) | Jackson Immuno Research Labs | Cat# 715-165-151; RRID:AB_2315777 | IF (1:500) |
| Sequence-based reagent | 18 S_F | Gomez-Sanchez et al., 2009 | PCR primers | CGGCTACCACATCCAAGGAA |
| Sequence-based reagent | 18 S_R | Gomez-Sanchez et al., 2009 | PCR primers | GCTGGAATTACCGCGGCT |
| Sequence-based reagent | Bdnf_F | Ma et al., 2016 | PCR primers | GGTATCCAAAGGCCAACTGA |
| Sequence-based reagent | Bdnf_R | Ma et al., 2016 | PCR primers | GCAGCCTTCCTTGGTGTAAC |
| Sequence-based reagent | Jun_F | Arthur-Farraj et al., 2012 | PCR primers | CCTTCTACGACGATGCCCTC |
| Sequence-based reagent | Jun_R | Arthur-Farraj et al., 2012 | PCR primers | GGTTCAAGGTCATGCTCTGTTT |
| Sequence-based reagent | Ednrb_F | NM_007904.4 | PCR primers | CTGGCTCTGGGAGACCTACT |
| Sequence-based reagent | Ednrb_R | NM_007904.4 | PCR primers | GGGCACCAGCTTACACATCT |
| Sequence-based reagent | Gdnf_F | Ma et al., 2016 | PCR primers | TCTCGAGCAGGTTCGAATGG |
| Sequence-based reagent | Gdnf_R | Ma et al., 2016 | PCR primers | AAGAACCGTCGCAAACTTTACC |
| Sequence-based reagent | Hdac4_F | NM_207225.2 | PCR primers | GCGAGCACAGAGGTGAAGAT |
| Sequence-based reagent | Hdac4_R | NM_207225.2 | PCR primers | CGCTGGAAATGCAGTGGTTC |
| Sequence-based reagent | Hdac5_F | NM_001077696.1 | PCR primers | GGGGTGGAGGTGGAGGTAG |
| Sequence-based reagent | Hdac5_R | NM_001077696.1 (20) | PCR primers | CCGTAGCGCAGGGTCCAT |
| Sequence-based reagent | Hdac7_F | NM_001204275.1 | PCR primers | AGGAGCAAGAACTTCGGCAA |
| Sequence-based reagent | Hdac7_R | NM_001204275.1 | PCR primers | ACTGTTCTCTCAAGGGCTGC |
| Sequence-based reagent | Hdac9_F | NM_001271386.1 | PCR primers | CCCCTATGGGAGATGTTGAG |
| Sequence-based reagent | Hdac9_R | NM_001271386.1 | PCR primers | CAATGCATCAAATCCAGCAG |
| Sequence-based reagent | Hmgcr_F | Gomez-Sanchez et al., 2009 | PCR primers | TGGATCGAAGGACGAGGAAAG |
| Sequence-based reagent | Hmgcr_R | Gomez-Sanchez et al., 2009 | PCR primers | GAATTACGTCAACCATAGCTTCCG |
| Sequence-based reagent | Krox-20_F | NM_010118.3 | PCR primers | ACCCCTGGATCTCCCGTATC |
| Sequence-based reagent | Krox-20_R | NM_010118.3 | PCR primers | CAGGGTACTGTGGGTCAATGG |
| Sequence-based reagent | Mbp_F | Gomez-Sanchez et al., 2009 | PCR primers | ATCCAAGTACCTGGCCACAG |
| Sequence-based reagent | Mbp_R | Gomez-Sanchez et al., 2009 | PCR primers | CCTGTCACCGCTAAAGAAGC |
| Sequence-based reagent | Mcam_F | NM_023061.2 | PCR primers | GAAACGGCTACCCCATTCCT |
| Sequence-based reagent | Mcam_R | NM_023061.2 | PCR primers | AGCCACTGGACTCGACAATC |
| Sequence-based reagent | Mef2a_F | NM_001033713.2 | PCR primers | AGTAGCGGAGACTCGGAATTG |
| Sequence-based reagent | Mef2a_R | NM_001033713.2 | PCR primers | ATGCATCGTACACAGCTCCT |
| Sequence-based reagent | Mef2c_F | Materna et al., 2019 | PCR primers | GTGCTGTGCGACTGTGAGAT |
| Sequence-based reagent | Mef2c_R | Materna et al., 2019 | PCR primers | TCTGAGTTTGTCCGGCTCTC |
| Sequence-based reagent | Mef2d_F | NM_001310587.1 | PCR primers | GATCTGAACAATGCCCAGCG |
| Sequence-based reagent | Mef2d_R | NM_001310587.1 | PCR primers | GGCAGCTGGTAATCTGTGTTG |
| Sequence-based reagent | Mitf_F | NM_001113198.1 | PCR primers | GGAGCTCACAGCGTGTATTT |
| Sequence-based reagent | Mitf_R | NM_001113198.1 | PCR primers | TCCTTAATGCGGTCGTTTATGT |
| Sequence-based reagent | Mpz_F | Gomez-Sanchez et al., 2009 | PCR primers | ACCAGACATAGT GGGCAAGACCTC |
| Sequence-based reagent | Mpz_R | Gomez-Sanchez et al., 2009 | PCR primers | AAGAGCAACAGC AGCAACAGCACC |
| Sequence-based reagent | Ngfr_F | Fontana et al., 2012 | PCR primers | TGATGGAGTCGGGCTAATGTC |
| Sequence-based reagent | Ngfr_R | Fontana et al., 2012 | PCR primers | AGATTCATCCCTCCACAAATGC |
| Sequence-based reagent | Olig1_F | Ma et al., 2016 | PCR primers | AGCGATGTAGTTGCTTGGGAT |
| Sequence-based reagent | Olig1_R | Ma et al., 2016 | PCR primers | CTGGCTCTAAACAGGTGGGAT |
| Sequence-based reagent | Pou3f1_F | NM_011141.2 | PCR primers | GAGCACTCGGACGAGGATG |
| Sequence-based reagent | Pou3f1_R | NM_011141.2 | PCR primers | TGATGCGTCGTTGCTTGAAC |
| Sequence-based reagent | Prx_F | NM_198048.2 | PCR primers | AGTGGCCAAGCTGAACATCC |
| Sequence-based reagent | Prx_R | NM_198048.2 | PCR primers | AGAACTCGACGTCAACAGGG |
| Sequence-based reagent | Runx2_F | NM_001146038.2 | PCR primers | GTCTTCCACACGGGGCAC |
| Sequence-based reagent | Runx2_R | NM_001146038.2 | PCR primers | GCCAGAGGCAGAAGTCAGAG |
| Sequence-based reagent | Sox2_F | Quintes et al., 2016 | PCR primers | TCCAAAAACTAATCACAACAATCG |
| Sequence-based reagent | Sox2_R | Quintes et al., 2016 | PCR primers | GAAGTGCAATTGGGATGAAAA |
| Sequence-based reagent | Sox10_F | NM_011437.1 | PCR primers | GAGCAAGCCGCACGTCAAGA |
| Sequence-based reagent | Sox10_R | NM_011437.1 | PCR primers | GTGGAGGTGAGGGTACTGGTC |
| Sequence-based reagent | Shh_F | Ma et al., 2016 | PCR primers | CAGCGACTTCCTCACCTTCCT |
| Sequence-based reagent | Shh_R | Ma et al., 2016 | PCR primers | AGCGTCTCGATCACGTAGAAGAC |
| Sequence-based reagent | Tyrp1_F | NM_031202.3 | PCR primers | CCGCTTTTCTCACATGGCAC |
| Sequence-based reagent | Tyrp1_R | NM_031202.3 | PCR primers | TCGCAGACGTTTTTCCCAGT |
| Sequence-based reagent | ChIP Hdac7 Promoter_F | ID_84582 | PCR primers | CCCTCCACAATGACCCTCCTT |
| Sequence-based reagent | ChIP Hdac7 Promoter_R | ID_84582 | PCR primers | GTGATCCGCTGTAATGCACTG |
| Sequence-based reagent | ChIP Hdac9 Promoter_F | ID_687001 | PCR primers | GCTGCAATCACTCGGCCAT |
| Sequence-based reagent | ChIP Hdac9 Promoter_R | ID_687001 | PCR primers | GCCCACAGGCACAGAAATAGA |
| Sequence-based reagent | ChIP Pou3f1 Promoter_F | ID_192110 | PCR primers | CAGAAGGAGAAGCGCATGAC |
| Sequence-based reagent | ChIP Pou3f1 Promoter_R | ID_192110 | PCR primers | CTCCCCAGGCGCATAAACG |
| Sequence-based reagent | Jun_FloxP_OE_F | Fazal et al., 2017 | PCR primers | TGGCACAGCTTAAGCAGAAA |
| Sequence-based reagent | Jun_FloxP_OE_R | Fazal et al., 2017 | PCR primers | GCAATATGGTGGAAAATAAC |
| Sequence-based reagent | Jun_FloxP_cKO_F | Arthur-Farraj et al., 2012 | PCR primers | CCGCTAGCACTC ACGTTGGTAGGC |
| Sequence-based reagent | Jun_FloxP_cKO_F | Arthur-Farraj et al., 2012 | PCR primers | CTCATACCAGTT CGCACAGGCGGC |
| Sequence-based reagent | Hdac4_FloxP_F | Gomis-Coloma et al., 2018 | PCR primers | ATCTGCCCACCAGAGTATGTG |
| Sequence-based reagent | Hdac4_FloxP_R | Gomis-Coloma et al., 2018 | PCR primers | CTTGTTGAGAAC AAACTCCTGCAGCT |
| Sequence-based reagent | Hdac5_FloxP_F | Gomis-Coloma et al., 2018 | PCR primers | CAAGGCCTTGTG CATGCTGGGCTGG |
| Sequence-based reagent | Hdac5_FloxP_R | Gomis-Coloma et al., 2018 | PCR primers | CTGCTCCCGTAG CGCAGGGTCCATG |
| Sequence-based reagent | Hdac5_FloxP_LacZ | Gomis-Coloma et al., 2018 | PCR primers | GCCCGTTTGA GGGGACGACG ACAGTATTCG |
| Sequence-based reagent | Hdac7_FloxP_F | Chang et al., 2006 | PCR primers | GTTGCAGGGTC AGCAGCGCAGGCTCTG |
| Sequence-based reagent | Hdac7_FloxP_R | Chang et al., 2006 | PCR primers | CCAGTGGACGAG CATTCTGGAGAAAGG |
| Sequence-based reagent | Mpz-Cre_F | Feltri et al., 1999 | PCR primers | CCACCACCTCT CTCCATTGCAC |
| Sequence-based reagent | Mpz-Cre_R | Feltri et al., 1999 | PCR primers | GCTGGCCCAA ATGTTGCTGG |
| Commercial assay or kit | QIAquick PCR Purification Kit | Qiagen | Cat# 28,104 | |
| Commercial assay or kit | NucleoSpin RNA, Mini kit for RNA purification | Macherey-Nagel | Cat# 740955.50 | |
| Commercial assay or kit | Luciferase Assay System | Promega | Cat# E1500 | |
| Commercial assay or kit | Beta-Glo Assay System | Promega | Cat# E4720 | |
| Commercial assay or kit | Pierce BCA Protein Assay Kit | Thermo Scientific | Cat# 23,225 | |
| Commercial assay or kit | ECL Prime Western Blotting Detection Reagent | Amersham | Cat# RPN223 | Highly sensitive chemiluminescent detection reagent for Western blotting |
| Commercial assay or kit | ECL Western Blotting Analysis System | Amersham | Cat# RPN2109 | Chemiluminescent detection reagent for Western blotting |
| Commercial assay or kit | Invitrogen Dynabeads Protein G | Life Technologies | Cat# 10,004D | Superparamagnetic beads with recombinant Protein G covalently coupled to surface for Immunoprecipitation |
| Commercial assay or kit | MasterMix qPCR ROx PyroTaq EvaGreen 5 x | CMB | Cat# 08-24-00001 | |
| Chemical compound, drug | Agar 100 Resin | Agar Scientific | Cat# R1043 | Concentration: various, see methods |
| Chemical compound, drug | Dodecenyl Succinic Anhydride - DDSA | Agar Scientific | Cat# R1051 | Concentration: various, see methods |
| Chemical compound, drug | Methyl Nadic Anhydride - MNA | Agar Scientific | Cat# R1082 | Concentration: various, see methods |
| Chemical compound, drug | Benzyldimethylamine - BDMA | Agar Scientific | Cat# R1062 | Concentration: various, see methods |
| Chemical compound, drug | Paraformaldehyde 16% solution, EM grade | Electron Microscopy Sciences | Cat# 15,710 | Concentration: 2% |
| Chemical compound, drug | Glutaraldehyde 25% solution, EM grade | Electron Microscopy Sciences | Cat# 16,220 | Concentration: 2.5% |
| Chemical compound, drug | Sodium Cacodylate Trihydrate | Electron Microscopy Sciences | Cat# 12,300 | Concentration: 0.1 M |
| Chemical compound, drug | Ethanol absolute | J.T.Baker | Cat# 8025 | Concentration: various, see methods |
| Chemical compound, drug | Propylene Oxide, ACS Reagent | Electron Microscopy Sciences | Cat# 20,401 | Concentration: various, see methods |
| Chemical compound, drug | Leibovitz's L-15 Medium | Company | Cat# 11570396 | Concentration: various, see methods |
| Chemical compound, drug | DMEM GlutaMAX Medium | Gibco | Cat# 11574516 | Concentration: various, see methods |
| Chemical compound, drug | Forskolin | Gibco | Cat# F6886 | Concentration: various, see methods |
| Chemical compound, drug | rhNRG1-beta1 | Sigma-Aldrich | Cat# RYD-396-HB-050 | Concentration: various, see methods |
| Chemical compound, drug | FBS - Fetal Bovine Serum | R&D Systems | Cat# 11550356 | Concentration: various, see methods |
| Chemical compound, drug | Penicillin-Streptomycin | Fisher | Cat# 11548876 | Concentration: various, see methods |
| Chemical compound, drug | DMEM Ham’s F12 Medium | Gibco | Cat# 11520396 | Concentration: various, see methods |
| Chemical compound, drug | Insulin-Transferrin-Selenium | Gibco | Cat# 41400-045 | Concentration: various, see methods |
| Chemical compound, drug | Putrescine | Gibco | Cat# P5780 | Concentration: various, see methods |
| Chemical compound, drug | Progesterone | Sigma-Aldrich | Cat# P0130 | Concentration: various, see methods |
| Chemical compound, drug | dbcAMP - Dibutyryl cAMP sodium salt | Sigma-Aldrich | Cat# D0627 | Concentration: various, see methods |
| Chemical compound, drug | LipoD293 DNA In Vitro Transfection Reagent | Sigma-Aldrich | Cat# SL100668 | Concentration: various, see methods |
| Chemical compound, drug | Mini Protease Inhibitor Cocktail | Roche | Cat# 11836153001 | Concentration: various, see methods |
| Chemical compound, drug | Phosphatase Inhibitor Mini Tablets | Fisher Scientific | Cat# 15691759 | Concentration: various, see methods |
| Chemical compound, drug | RNase A | Fisher Scientific | Cat# 10618703 | Concentration: various, see methods |
| Chemical compound, drug | Proteinasa K | Sigma-Aldrich | Cat# 3115836001 | Concentration: various, see methods |
| Chemical compound, drug | TRI Reagent | Sigma-Aldrich | Cat# T9424 | Concentration: various, see methods |
| Chemical compound, drug | Chloroform | Sigma-Aldrich | Cat# 319,988 | Concentration: various, see methods |
| Chemical compound, drug | DNaseI RNase Free | Thermo Fisher Scientific | Cat# EN0521 | Concentration: various, see methods |
| Chemical compound, drug | Deoxynucleotide Mix | Sigma-Aldrich | Cat# D7295 | Concentration: various, see methods |
| Chemical compound, drug | Random Primers | Invitrogen | Cat# 48190-011 | Concentration: various, see methods |
| Chemical compound, drug | DTT | Invitrogen | Cat# Y00147 | Concentration: various, see methods |
| Chemical compound, drug | RNaseOUT | Invitrogen | Cat# 10777-019 | Concentration: various, see methods |
| Chemical compound, drug | RevertAid Reverse Transcriptase | Thermo Fisher Scientific | Cat# EP0441 | Concentration: various, see methods |
| Software, algorithm | GraphPad Prism 9.0.0 | GraphPad Prism | RRID:SCR_002798 | |
| Software, algorithm | ImageJ | Open Source | RRID:SCR_003070 | |
| Software, algorithm | QuantStudio 3 Real-Time PCR Systems Software | Applied Biosystems | RRID:SCR_018712 | |
| Other | QuantStudio 3 Real-Time PCR Systems | Applied Biosystems | Cat# A28131 | Real Time PCR thermal cycler |
| Other | Biorruptor Pico sonication device | Diagenode | Cat# B01060010 | Sonication device |
| Other | Bullet Blender Homogenizer BBX24-CE | Next Advance | Cat# BBX24-CE | Tissue Homogenizer |
| Other | Zirconium Oxide Beads | Next Advance | Cat# ZrOB05 | For the homogenization of medium-tough tissue and cells |
| Other | Microplate Reader EZ Read 400 | Biochrom | Cat# 12694795 | 96-well plate reader for Pierce BCA protein assay kit |
| Other | Nitrocellulose Membrane | Amersham Hybond ECL | Cat# RPN203D | Protein blotting membrane (pore size: 0.45 mm) |
| Other | Amersham Imager 600 | Amersham | Cat# Amersham Imager 600 | Western blot (chemiluminescence) developer |
| Other | 4',6-diamidino-2-phenylindole (DAPI stain) | Thermo Fisher | Cat# D1306; RRID:AB_2629482 | Blue-fluorescent nucleic acid stain IF (1:1000) |
Additional files
-
Source data 1
Graphs source data.
- https://cdn.elifesciences.org/articles/72917/elife-72917-data1-v2.docx
-
Source data 2
RNAseq source data.
- https://cdn.elifesciences.org/articles/72917/elife-72917-data2-v2.xlsx
-
Source data 3
ChIPseq source data.
- https://cdn.elifesciences.org/articles/72917/elife-72917-data3-v2.xlsx
-
Source data 4
Western blot source data.
- https://cdn.elifesciences.org/articles/72917/elife-72917-data4-v2.pdf
-
Reporting standard 1
ARRIVE guidelines checklist.
- https://cdn.elifesciences.org/articles/72917/elife-72917-repstand1-v2.pdf





