Innexin function dictates the spatial relationship between distal somatic cells in the Caenorhabditis elegans gonad without impacting the germline stem cell pool
Figures
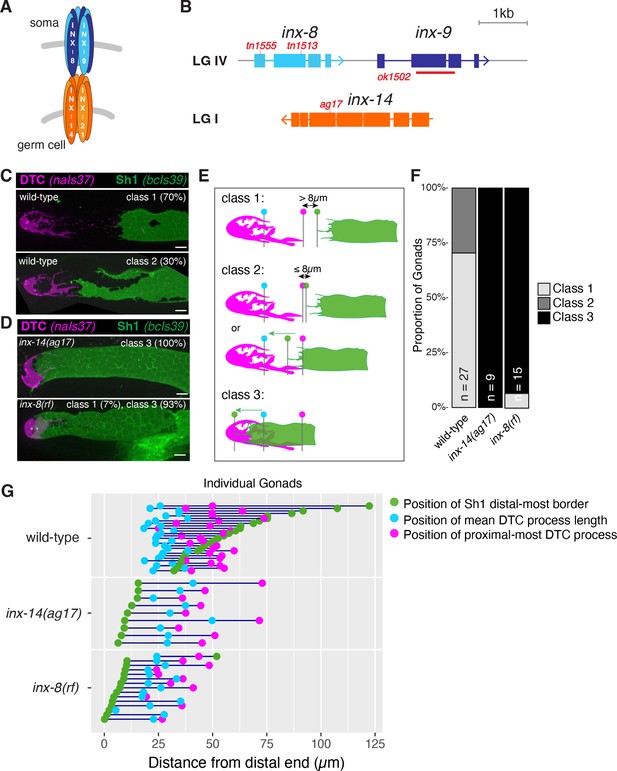
Germline and somatic gonad innexins are required for proper somatic gonad architecture.
(A) Schematic of paired somatic and germline octameric hemichannels. (B) Schematic diagram of the inx-8 inx-9 locus and the inx-14 locus, with relevant mutations indicated in red. (C) Fluorescent confocal maximum projection images of distal gonads in live worms. Distal tip cell (DTC) marked in magenta (naIs37[lag-2p::mCherry-PH]) and sheath pair 1 (Sh1) marked in green (bcIs39[lim-7p::CED-1::GFP]). Top: strain bearing markers only, denoted “wild-type”, representative of phenotypic Classes 1 and 2. (D) inx-14(ag17) and inx-8(tn1513tn1555) inx-9(ok1502), denoted ‘inx-8(rf)’ after Starich et al., 2020, representative of Class 3. (E) Diagram of DTC–Sh1 relationship in distal end of a typical wild-type gonad (DTC magenta, Sh1 green) for phenotypic Classes 1–3. Class 1: greater than 8 µm (two cell diameters) separates the proximal most extent of the DTC (vertical line topped with magenta dot) and the distal most border of Sh1 (vertical line topped with green dot) as measured in slice-by-slice analysis of Z series; Class 2: ≤8 µm separates DTC and Sh1 extensions or interdigitation of DTC and Sh1 is seen up to the position of the average DTC process length (vertical line topped with blue dot); Class 3: distal position of Sh1 is distal to the average DTC process length. A diagram of all measurements taken for live fluorescent images is shown in Figure 1—figure supplement 1. (F) The proportion of gonads examined that fall into the phenotypic classes indicated. (G) Plot showing the distance between the average DTC process length (blue dots), the longest DTC process (magenta dots) and the distal-most border of Sh1 (green dots); each trio of dots joined by a dark blue line represents the data for a single gonad, analyzed as a confocal maximum projection. Scale bars are 10 µm. Strains used are DG5020, DG5026, and DG5029; n for each is indicated in panel F. See Supplementary file 1 for complete genotypes.
-
Figure 1—source data 1
Source data for Figure 1C–D and Figure 1F–G.
- https://cdn.elifesciences.org/articles/74955/elife-74955-fig1-data1-v1.xlsx
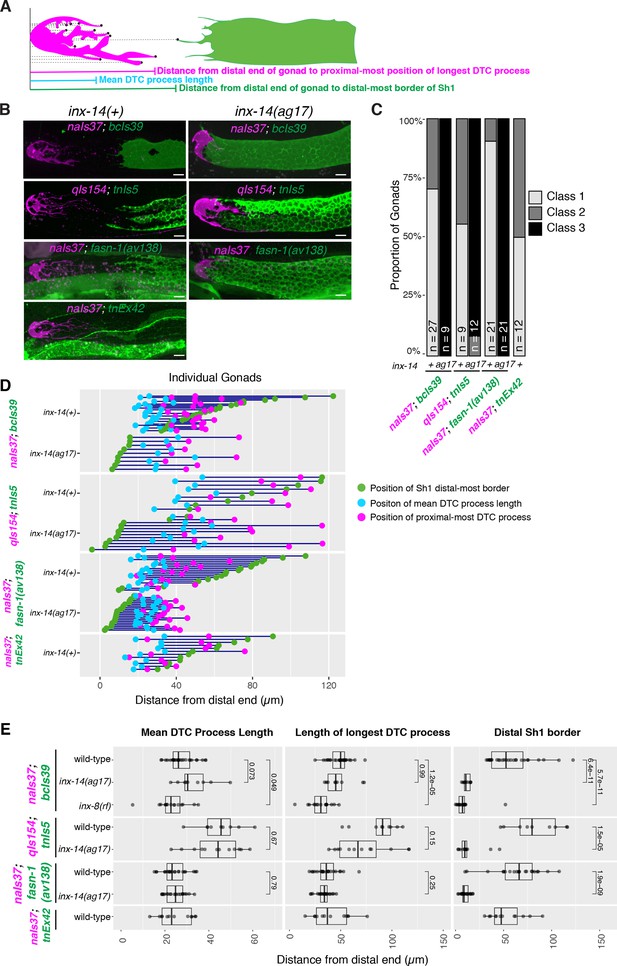
Consistent trends in Sh1 and DTC positions are observed with multiple markers.
(A) Schematic diagram of DTC and Sh1 parameters measured. Each black dot represents a point in a maximum projection Z-stack for which the distance from the distal end was measured in microns. (B) Fluorescent micrographs of gonads in live worms bearing indicated markers that label DTC (magenta lettering) or Sh1 (green lettering), in wild-type and inx-14(ag17) mutant backgrounds, as indicated: naIs37, bcIs39, qIs154, and tnIs5 are integrated transgenes; tnEx42 carries a rescuing acy-4p::gfp fusion on an extrachromosomal array; and fasn-1(av138[fasn-1::gfp]) was tagged at the endogenous locus using CRISPR-Cas9 genome editing (see Supplementary file 1 for full genotypes). The fasn-1 and acy-4 fusions also drive expression in non-gonadal tissues. (C) The proportion of gonads examined that fall into the phenotypic classes indicated. (D) Plot showing the distance between the average DTC process length (blue dots), the longest DTC process (magenta dots) and the distal-most border of Sh1 (green dots); each trio of dots joined by a dark blue line represents the data for a single gonad. (E) Plots showing quantitative measurement of parameters diagrammed in (A). Although the behavior of the DTCs marked by different markers is variable, and DTCs marked with qIs154 show significantly longer DTC processes, the behavior of Sh1 in the presence of either marker shows significantly consistent changes across mutant genotypes. All P values result from Student’s t-test. Scale bars are 10 µm. Strains used were DG5020, DG4959, DG4977, DG5026, DG5320, DG5367, DG5310; n for each is indicated in panel C. See Supplementary file 1 for complete genotypes.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1B–E.
- https://cdn.elifesciences.org/articles/74955/elife-74955-fig1-figsupp1-data1-v1.xlsx
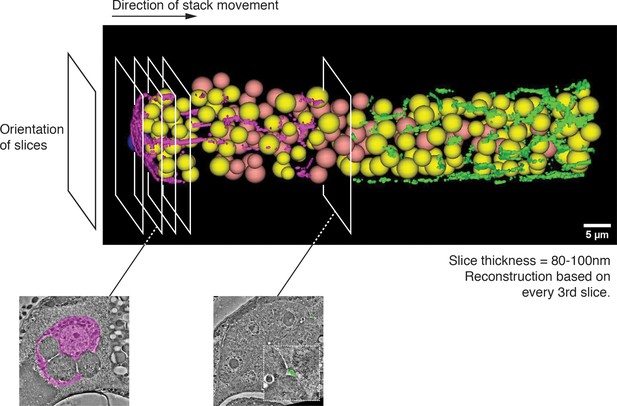
Snapshot of transmission electron micrograph (TEM) reconstruction and guide to Videos 4 and 5.
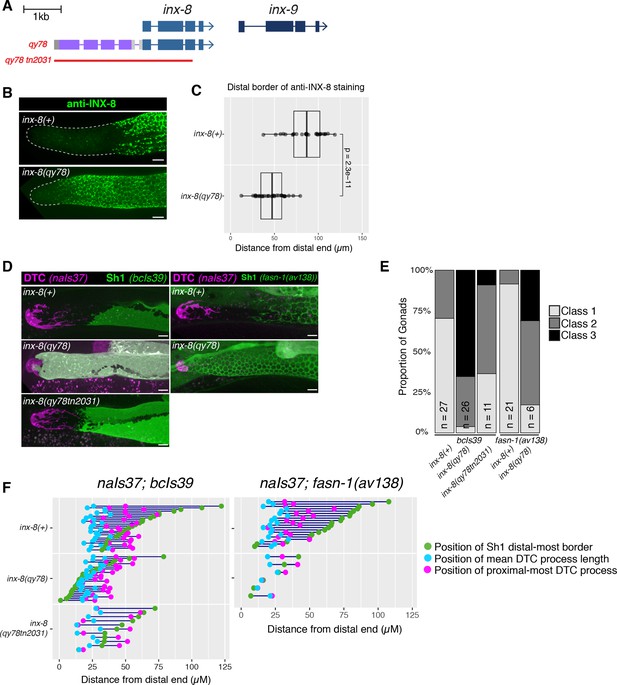
N-terminal fusion of mKate2 to INX-8 generates an INX-8 protein that alters somatic gonad morphology.
(A) Schematic diagram showing the genetic manipulations used in this section. inx-8(qy78[mKate2::inx-8]) was created by placing mKate2 in-frame with the N-terminus of INX-8 (Gordon et al., 2020). inx-8(qy78tn2031) was created by deleting the inx-8 coding region and mKate2 moiety in the inx-8(qy78) background. (B) Representative distal gonads stained with anti-INX-8 antibody; Top: N2 wild type, Bottom: DG5063 inx-8(qy78[mKate2::inx-8]). See legend to Figure 2—figure supplement 1 for further details. (C) Dot plot with overlaid quantile box plots showing the distance from the distal end of the gonad to the Sh1 distal border for each genotype. Each dot represents a single gonad of that genotype. p value was calculated using Student’s t-test. (D) Fluorescent micrographs of live animals with the DTC marked by naIs37[lag-2p::mCherry-PH] and Sh1 marked by bcIs39[lim-7p::ced-1::gfp]. Top: wild-type with markers only. Middle: inx-8(qy78[mKate2::inx-8]). Bottom: inx-8(qy78tn2031). (E) The proportion of gonads examined that fall into the phenotypic classes indicated. (F) Quantitative dot-plots as in Figure 1; data for wild-type (marker only) strain in E and F is the same as in Figure 1. Each pair of dots connected by a line represents data from a single gonad. Scale bar 10 µm. Strains used were N2, DG5063, DG5020, DG5131, DG5229, DG5320, DG5378; n for each is indicated in panel E. See Supplementary file 1 for complete genotypes.
-
Figure 2—source data 1
Source data for Figure 2B and C.
- https://cdn.elifesciences.org/articles/74955/elife-74955-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Source data for Figure 2D–F.
- https://cdn.elifesciences.org/articles/74955/elife-74955-fig2-data2-v1.xlsx
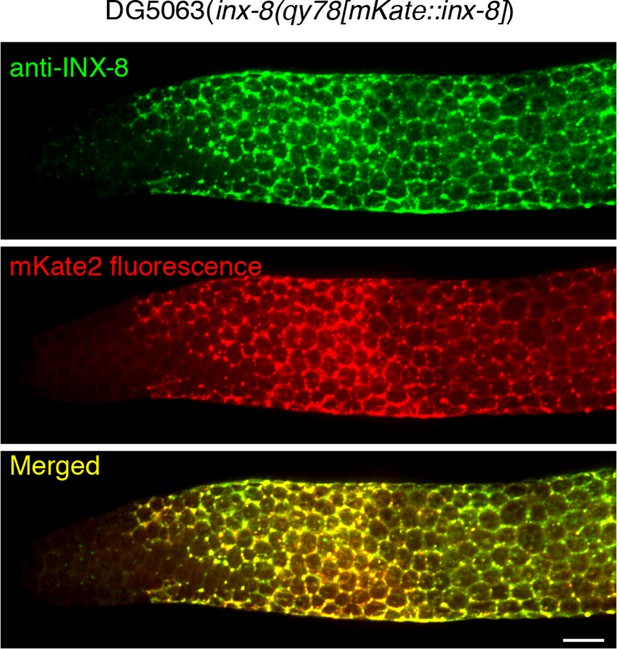
INX-8 antibody staining overlaps with mKate2 fluorescence in inx-8(qy78[mKate2::inx-8)] gonads.
Shown are fluorescence confocal maximum projections of surface images (~7 µm depth) of a DG5063 inx-8qy78[mKate2::INX-8] dissected and fixed gonad, with INX-8 visualized by anti-INX-8 antibody staining (top, green), endogenous mKate2 fluorescence (middle, red), and merged (bottom, yellow). The distal end of the gonad is on the left. In all gonads examined (n=32), overlap was complete. Scale bars are 10 µm. The INX-8 distribution pattern in wild type appears less punctate compared to a previous report (Starich et al., 2014). We attribute this to several differences in imaging. (1) Images in Starich et al., 2014 were obtained on a compound microscope, whereas the images shown here were acquired by confocal microscopy. (2) Except for the progenitor zone region (see point 3 below), images in Starich et al., 2014 were captured to present a surface view, while images presented here are maximum projections of confocal stacks, as described above. This stack goes from the gonad surface down through to the surface germ cell nuclei, which results in the appearance of increased signal from junctions residing between intercalating Sh1 invaginations and germ cells, as well as from INX-8 trafficking through the secretory pathway. This gives the appearance of a more honeycomb antibody staining pattern than Starich et al., 2014. Unlike the surface view image from the compound microscope, a projection of confocal images is necessary to capture both Sh1 and the DAPI- stained germ cell nuclei that served as a reference (DAPI is not shown). (3) For the progenitor zone (labelled M for mitotic region in the 2014 paper), the image in Figure 1 of Starich et al., 2014 shows a slightly more interior view (at the level of the rachis), revealing sparse punctate staining near the distal tip of the gonad. In this study, DTC staining is less apparent since the distal-most region is also maximum projection confocal stacks only from the gonad surface to the surface germ cell nuclei. Importantly, when viewing more interior planes of the Z-stack, sparse punctate staining is observed at positions consistent with DTC-rachis contact, as described in Figure 6 of Starich et al., 2014. (4) Finally, fixation conditions in this study were slightly different than Starich et al., 2014, so as to preserve mKate2 fluorescence.
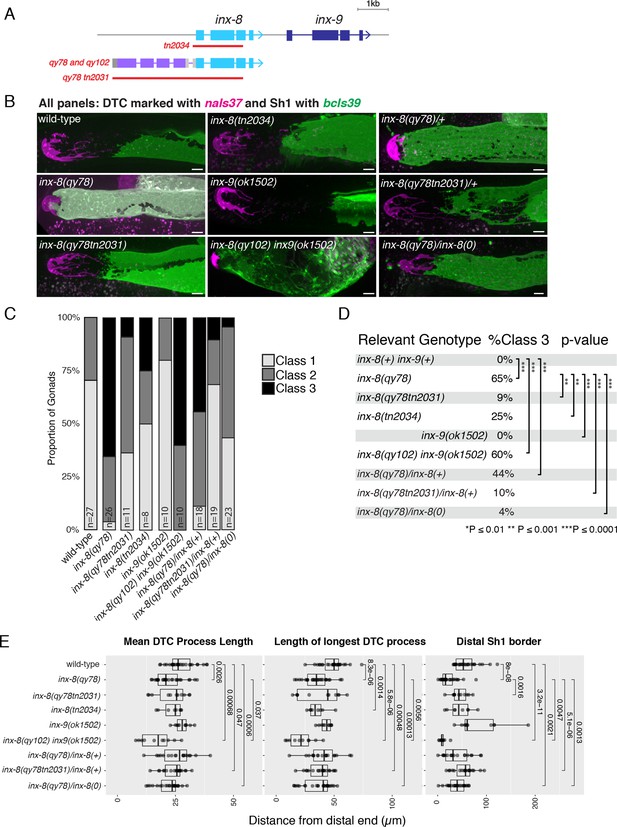
The inx-8(qy78[mKate2::inx-8]) allele behaves as a dosage-sensitive antimorph.
(A) Schematic diagram showing additional relevant alleles at the inx-8 and inx-9 loci. (B–E) Relevant genotypes are given for inx-8 and inx-9 only. All strains also contain naIs37 to mark the DTC and bcIs39 to mark Sh1. inx-8(qy78[mKate2::inx-8]) and inx-8(qy102[mKate2::inx-8]) are abbreviated as inx-8(qy78) and inx-8(qy102) to aid comparisons between genotypes. (B) Representative fluorescent micrographs of live worms carrying each relevant allele, as indicated (images for inx-8(+), inx-8(qy78[mKate2::inx-8]) and inx-8(qy102tn2031) are the same as in Figure 2B, included here for comparison). (C) Stacked histogram showing the frequency of each of the phenotypic classes described in Figure 1, for each genotype, as indicated. (D) Table showing frequency of Class 3 gonads for each genotype, with indicated pairwise significance levels derived from a Fisher’s Exact Test. (E) Quantitative plots showing changes to DTC morphology and Sh1 position. All P values result from Student’s t-test. Scale bars are 10 µm. Strains used were DG5020, DG5131, DG5229, DG5232, DG5027, DG5133, DG5346, DG5347, DG5366; n for each is indicated in panel C. See Supplementary file 1 for complete genotypes.
-
Figure 2—figure supplement 2—source data 1
Source data for Figure 2—figure supplement 2B–E.
- https://cdn.elifesciences.org/articles/74955/elife-74955-fig2-figsupp2-data1-v1.xlsx
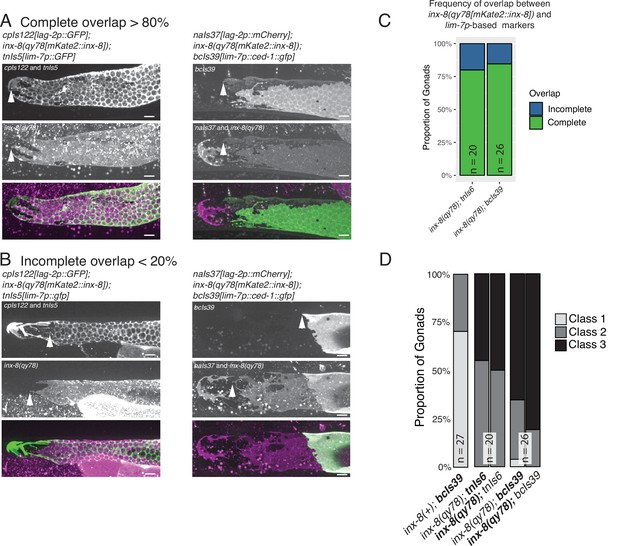
Direct comparison of inx-8(qy78[mKate2::inx-8]) and lim-7p-driven transgenes as markers for Sh1.
(A) Individual gonads carrying different Sh1 markers in addition to inx-8(qy78[mKate2::inx-8]). Left, top to bottom: Individual channels and merged image of a single worm co-expressing the DTC marker cpIs122[lag-2p::gfp], inx-8(qy78[mKate2::inx-8]) and the sheath cell marker tnIs5[lim-7p::gfp]. Right, top to bottom: Individual channels and merged image of a single worm co-expressing the DTC marker naIs37[lag-2p::mCherry-PH], inx-8(qy78[mKate2::inx-8]) and the sheath cell marker bcIs39[lim-7p::ced-1::gfp]. (B) Two individual gonads of the same genotypes as in (A), showing incomplete overlap between mKate2::INX-8 and the respective GFP sheath cell markers. White arrowheads mark the most distal extent of each marker in the single-channel panels. (C and D): inx-8(qy78)[mKate2::inx-8] is abbreviated as ‘inx-8(qy78)’. (C) Frequency of overlap. Of 46 gonads examined, 39 showed perfect overlap as shown in these images. (D) Proportion of gonads in phenotypic Classes 1–3 as scored by mKate2 or as scored by GFP expression, as indicated by bold in the genotype. Scale bars are 10 µm. Strains used were DG5020, DG5131, KLG006; n for each is indicated in panels C and D. See Supplementary file 1 for complete genotypes.
-
Figure 2—figure supplement 3—source data 1
Source data for Figure 2—figure supplement 3.
- https://cdn.elifesciences.org/articles/74955/elife-74955-fig2-figsupp3-data1-v1.xlsx
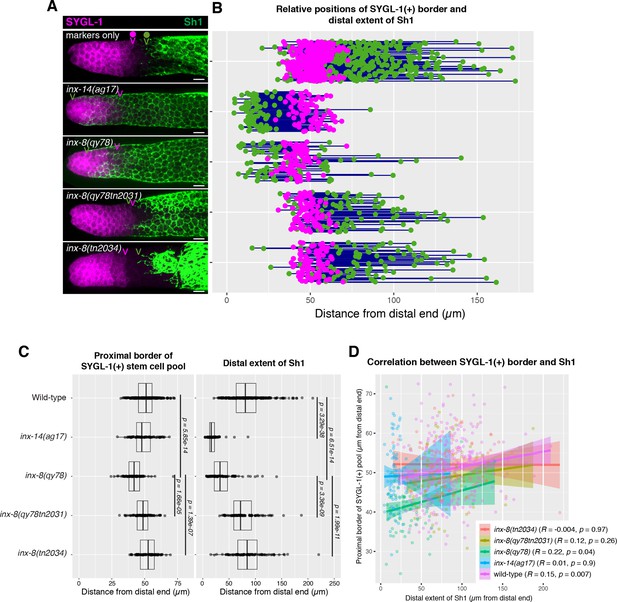
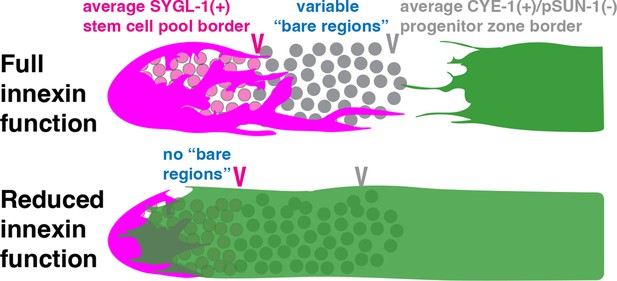
The position of the proximal border of the SYGL-1-positive stem cell pool does not correlate with the position of Sh1.
(A) Fluorescence confocal maximum projection of surface images (~7 µm depth) of fixed, dissected gonads showing the SYGL-1-positive stem cell pool marked in magenta and the sheath cell marked in green. Magenta caret represents the location of the proximal border of the SYGL-1-positive stem cell pool. Green caret represents the distal edge of Sh1. (B) Quantitative graph with magenta dots representing the proximal border of SYGL-1::OLLAS expression and green dots representing the distal reach of Sh1. Each pair of dots connected by a line represents the data for a single gonad. (C) Dot plot with overlaid box plots showing the distance and quantiles of the SYGL-1-positive stem cell pool and distal extent of Sh1 for each genotype. Each dot represents a single specimen of that genotype. p values were calculated using Student’s t-test. (D) Scatterplot showing lack of correlation between the proximal extent of SYGL-1 expression and the distal reach of Sh1. Scale bars are 10 µm. Strains used, top to bottom as indicated on figure panels A and C, and (n) number of gonads are DG5136 (320), DG5150 (83), DG5181 (88), DG5248 (86), and DG5249 (88). See Supplementary file 1 for complete genotypes.
-
Figure 3—source data 1
Source data for Figure 3 and Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/74955/elife-74955-fig3-data1-v1.xlsx
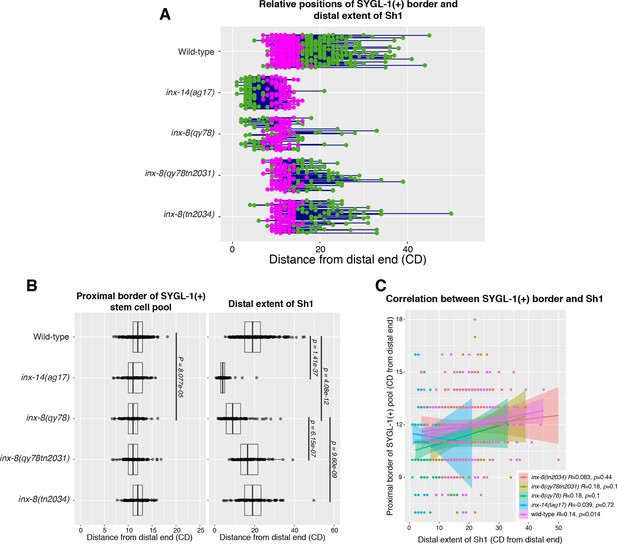
The position of the proximal border of the SYGL-1-positive stem cell pool does not correlate with the position of Sh1 when data are shown in cell diameters.
(A) Quantitative graph with magenta dots representing the proximal extent of SYGL-1::OLLAS expression and green dots representing the distal reach of Sh1. Each pair of magenta and green dots connected by a line represents the data for a single gonad. (B) Dot plot with overlaid box plots showing the distance and quantiles of the SYGL-1-positive stem cell pool and distal extent of Sh1 for each genotype, in cell diameters from the distal end. Each dot represents a single specimen of that genotype. p values were calculated using Student’s t-test. (C) Scatterplot showing lack of correlation between the proximal extent of SYGL-1 expression and the distal reach of Sh1. CD: cell diameters. Strains used and n are listed in Figure 3.
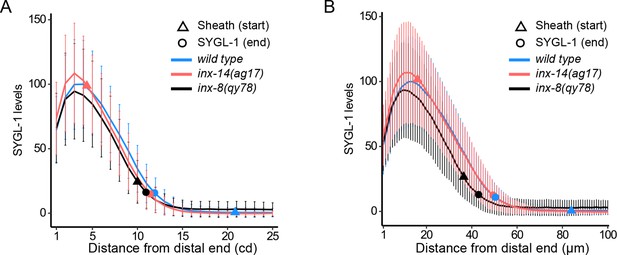
Proximal end of the SYGL-1 zone assessed visually and from protein levels.
SYGL-1 (3xOLLAS::SYGL-1) levels were measured from the distal tip of the gonad to (A) 25 CD and (B) 100 µmicrons proximal, in dissected and stained gonads from 1 day adults of the indicated genotypes. The visually assessed SYGL-1 border and the distal extent of Sh1 are marked. SYGL-1 protein levels from fluorescence-intensity measurements are normalized by setting the peak levels in wild type to 100. The SYGL-1 quantitation of the indicated genotype is carried out for the gonads in Figure 3B. The small differences observed in SYGL-1 peak levels between wild type and inx-14(ag17) or inx-8(qy78[mKate2::inx-8]) are not statistically significant (Student’s t-test). Triangle, distal most position (start) of Sh1; circle, proximal position of SYGL-1 staining; blue, wild type; red inx-14(ag17); black inx-8(qy78[mKate2::inx-8]). Curved plot lines, mean; vertical bars, mean ± SD. All three strains have sygl-1(q983) naIs37 I; bcIs39 V in the background along with the indicate inx-x genotype. Strains used are DG5136, DG5150, and DG5181. See Supplementary file 1 for complete genotypes.
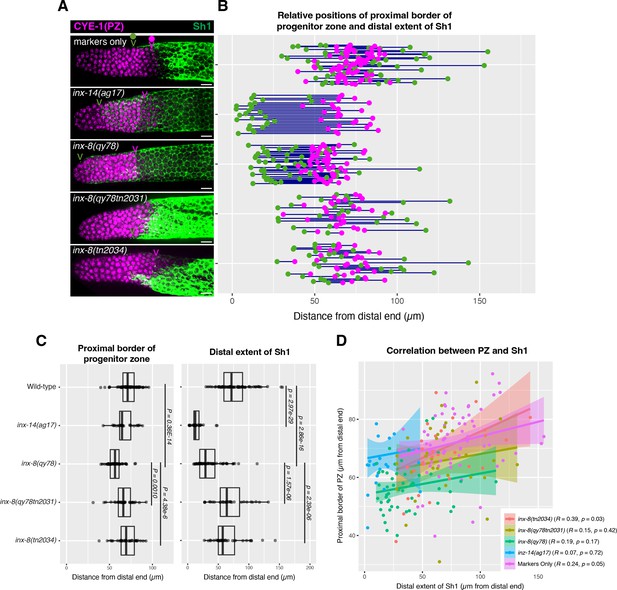
The position of the proximal border of the progenitor zone does not correlate with the position of Sh1.
(A) Fluorescence confocal maximum projection of surface images (~7 µm depth) of fixed, dissected gonads showing the progenitor pool marked in magenta and the sheath cell marked in green. All measurements were made by examining optical sections through the entire depth of the gonad. Magenta caret represents the location of the proximal border of the CYE-1-positive, pSUN-1-negative progenitor pool (pSUN-1 staining not shown; see Materials and methods). Green caret represents the distal edge of Sh1. (B) Quantitative graph with magenta dots representing the proximal extent of the CYE-1 staining and green dots representing the distal reach of Sh1. Each pair of magenta and green dots connected by a line represents the data for a single gonad. (C) Dot plot with overlaid box plots showing the distance and quantiles of the progenitor pool and distal extent of Sh1 for each genotype. Each dot represents a single specimen of that genotype. p values were calculated using Student’s t-test. (D) Scatterplot showing lack of correlation between the proximal PZ border and Sh1 position. Scale bars are 10 µm. Strains used, top to bottom as indicated on figure panels A and C, and (n) number of gonads are DG5136 (72), DG5150 (26), DG5181 (52), DG5248 (30), and DG5249 (32). See Supplementary file 1 for complete genotypes.
-
Figure 4—source data 1
Source data for Figure 4 and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/74955/elife-74955-fig4-data1-v1.xlsx
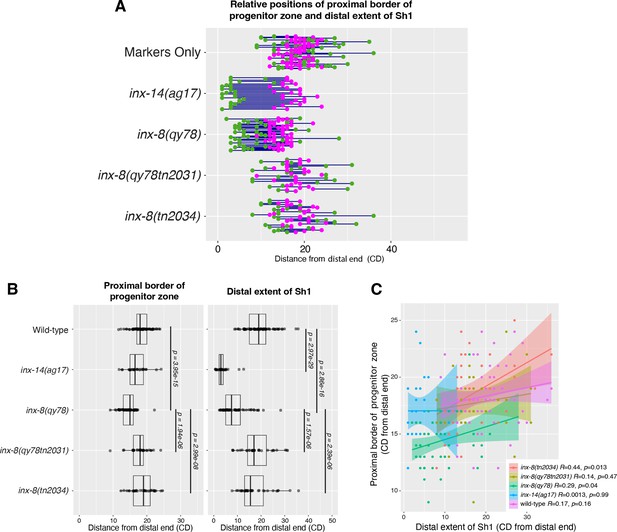
The position of the proximal border of the progenitor zone does not correlate with the position of Sh1 when data are shown in cell diameters.
(A) Quantitative graph with magenta dots representing the location of the proximal border of the CYE-1-positive, pSUN-1-negative progenitor pool and green dots representing the proximal extent of and green dots representing the distal reach of Sh1. Each pair of magenta and green dots connected by a line represents the data for a single gonad. (B) Dot plot with overlaid box plots showing the distance and quantiles of the CYE-1-positive progenitor pool and distal extent of Sh1 for each genotype, in cell diameters from the distal end. Each dot represents a single specimen of that genotype. p values were calculated using Student’s t-test. (C) Scatterplot showing lack of correlation between the proximal extent of progenitor zone and the distal reach of Sh1. CD: cell diameters. Strains used and n are listed in Figure 4.
Videos
Confocal stack and 3D rendering of representative Class 1 gonad.
Strain DG5020.
Confocal stack and 3D rendering of representative Class 2 gonad.
Strain DG5020.
Confocal stack and 3D rendering of representative Class 3 gonad.
Strain DG5026.
Serial thin section movie of every third section from the DTC to the distal extensions of Sh1 in a young adult hermaphrodite posterior gonad arm.
Germ cell nuclei are marked in yellow if the corresponding germ cells receive direct contact from a DTC or Sh1 process, or in salmon if they do not. The soma and processes of the DTC are filled in pink. Filopodial extensions of the somatic sheath are filled in green. Individual images are composed from multi-panel montages (mostly 3x4 montages). Scale bar is 3 µm.
Model of the distal gonad.
The DTC, its processes and fragments are in pink. Nearest sheath cell bodies lie out of frame proximally, but their filopodial extensions are shown in green, extending towards the DTC. Germ cell nuclei are modelled as spheres; the nuclei of those germ cells that do not come in contact with either DTC or Sh1 are colored in salmon, while germ cells with direct contact are shown in yellow. The true volume of each germ cell is larger than is shown here because each sphere is centered only on the cell nucleus. Scale bar is 5 µm.
Tables
Brood sizes and embryonic lethality measurements for selected strains.
| Strain | Relevant genotype* | Brood (n)† | %Lethality (n) ‡ | Sh1 mispositioned§ |
|---|---|---|---|---|
| N2 | Wild type | 298.6±42.1 (24) | 0.4±0.4 (7194) | no |
| DG5063 | inx-8(qy78) ¶ | 221.6±23.6 (25) | 9.0±4.6 (6087) | yes |
| NK2571 | inx-8(qy78); cpIs122 | 212.8±27.5 (23) | 6.2±3.0 (5217) | (yes**) |
| DG5059 | inx-9(ok1502) | 322.7±33.0 (25) | 0.2±0.3 (8081) | (no**) |
| DG5064 | inx-8(qy102) inx-9(ok1502)†† | 211.1±47.2 (25) | 1.1±1.1 (5327) | (yes**) |
| NK2576 | inx-8(qy102) inx-9(ok1502); cpIs122 | 209.5±52.7 (23) | 2.3±1.2 (4928) | (yes**) |
| DG5250 | inx-8(qy78tn2031) ‡ ‡ | 260.0±25.9 (25) | 0.1±0.2 (6504) | (no**) |
| DG5251 | inx-8(tn2034) ‡ ‡ | 301.6±37.8 (25) | 0.4±0.4 (7570) | (no**) |
| DG5270 | inx-14(ag17) | 253.2±28.1 (25) | 0.4±0.6 (6356) | (yes**) |
| DG5070 | inx-14(ag17); inx-8(qy78) | 106.6±19.4 (25) | 23.7±8.0 (3510) | n.d. |
| DG5380 | bcIs39 | 282.6±34.2 (25) | 0.4±0.5 (7091) | n.d. |
| DG5020 | naIs37; bcIs39 | 237.5±46.5 (24) | 1.0±1.4 (5753) | no |
| DG5320 | fasn-1(av138::gfp) naIs37 | 342.4±30.3 (25) | 0.4±0.4 (8591) | no |
| DG5367 | inx-14(ag17) fasn-1(av138::gfp) naIs37 | 256.0±26.6 (23) | 0.2±0.2 (5897) | yes |
| DG5378 | fasn-1(av138::gfp) naIs37; inx-8(qy78) | 245.4±43.7 (23) | 5.3±3.2 (5947) | yes |
-
*
Full genotypes in Supplementary file 1; genotypes inx-8(qy78[mKate2::inx-8]) and inx-8(qy102[mKate2::inx-8]) are abbreviated as “inx-8(qy78)” and “inx-8(qy102)” and fasn-1(av138[fasn-1::GFP]) as fasn-1(av138::gfp).
-
†
Brood size ± SD, n = number of broods counted.
-
‡
Percent lethality ± SD, n = number of embryos scored.
-
§
-
¶
Derived from NK2571 inx-8(qy78); cpIs122 [lag-2p::mNeonGreen::plcdeltaPH] (Gordon et al., 2020).
-
**
-
††
Derived from NK2576 inx-8(qy102) inx-9(ok1502); cpIs122 (Gordon et al., 2020).
-
‡ ‡
Equivalent inx-8(null) CRISPR-Cas9 deletions generated in inx-8(qy78[mKate2::inx-8]) or in wild-type genetic backgrounds.
-
Table 1—source data 1
Brood size and embryonic lethality data for Table 1.
- https://cdn.elifesciences.org/articles/74955/elife-74955-table1-data1-v1.xlsx
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | mouse monoclonal anti-CYE-1 | Brodigan et al., 2003 | (1:100) | |
| Antibody | rat monoclonal anti-OLLAS | Shin et al., 2017 | (1:2000) | |
| Antibody | guinea pig polyclonal anti-SUN-1 S8-Pi | Mohammad et al., 2018 | (1:1000) | |
| Antibody | rabbit polyclonal anti-GFP | from Swathi Arur, MD Anderson Cancer Center; Lopez et al., 2013 | (1:200) | |
| Antibody | rabbit polyclonal anti-INX-8 | Starich et al., 2014 | (1:25) | |
| sequence-based reagent | AF-ZF-827 | Arribere et al., 2014 | CACTTGAACTTCAATA CGGCAAGATGAGAAT GACTGGAAACCGT ACCGCATGCGGTG CCTATGGTAGCGGA GCTTCACATGGCTTCAG ACCAACAGCCTAT Contact DG for more information | |
| sequence-based reagent | inx8_delta.F | this work | CCTTCGACCTGATTT CCCCTTCTTCTAATG Contact DG for more information | |
| sequence-based reagent | inx8_delta.R | this work | CTATTGCTTTCCGTT CTTCAAGATGTTGTTG Contact DG for more information | |
| sequence-based reagent | inx8_RPR | this work | GGTGGCCAATAAA AATGCTTTTCTTTTT GCTTTTCTCTATCTA CTTCCGTTCCGCCCC GGAGGTTGCCGTGG AGATGTACAGCGAC TTTTTAGTAAGTCTT TTCAAC Contact DG for more information | |
| sequence-based reagent | inx8_sgRNA1.F | this work | TCTTGAGTGACTTGG TAGCATCGG Contact DG for more information | |
| sequence-based reagent | inx8_sgRNA1.R | this work | AAACCCGATGCTACC AAGTCACTC Contact DG for more information | |
| Recombinant DNA reagent (plasmid) | inx8_us_sgRNA1 | this work | available upon request Contact DG for more information | |
| sequence-based reagent | inx8_us_sgRNA1.F | this work | TCTTGTGGAAAACAG AGGAATGGG Contact DG for more information | |
| sequence-based reagent | inx8_us_sgRNA1.R | this work | AAACCCCATTCCTCT GTTTTCCAC Contact DG for more information | |
| sequence-based reagent | inx-14delF | this work | GATACGACGTGAGCA ATGGAACGTC Contact DG for more information | |
| sequence-based reagent | inx-14delR | this work | CTTGGACTTGAAGT GAGAGTTGGAG Contact DG for more information | |
| sequence-based reagent | sygl1-F | this work | ATCATCGAACCA TTGTCATCACGC Contact DG for more information | |
| sequence-based reagent | sygl1-R | this work | TTTGCCTTGATCTC CAAGTGTTGC Contact DG for more information | |
| Genetic reagent (C. elegans) | AG400 | Starich et al., 2020 | fasn-1(av138[fasn-1::gfp]) I | |
| Genetic reagent (C. elegans) | CB190 | Brenner, 1974 | unc-54(e190) I | |
| Genetic reagent (C. elegans) | CB1282 | Hosono et al., 1982 | dpy-20(e1282) IV | |
| Genetic reagent (C. elegans) | DG2506 | Govindan et al., 2009 | acy-4(ok1806) V; tnEx42[acy-4::gfp +rol-6(su1006)] | |
| Genetic reagent (C. elegans) | DG4959 | (this work) | qIs154[lag-2p::MYR::tdTomato +ttx-3p::gfp] V; tnIs5[lim-7p::gfp +rol-6(su1006)] X | Contact DG for more information |
| Genetic reagent (C. elegans) | DG4977 | (this work) | inx-14(ag17) I; qIs154[lag-2p::MYR::tdTomato +ttx-3p::gfp] V; tnIs5[lim-7p::gfp +rol-6(su1006)] X | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5020 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5026 | (this work) | inx-14(ag17) naIs37[lag-2p::mCherry::PH +unc-119(+)] I; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5027 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-9(ok1502) IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5029 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(tn1513tn1555) inx-9(ok1502) IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5059 | (this work) | inx-9(ok1502) IV | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5063 | (this work) | inx-8(qy78[mKate2::inx-8]) IV | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5064 | (this work) | inx-8(qy102(mKate2::inx-8)) inx-9(ok1502) IV | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5070 | (this work) | inx-14(ag17) I; inx-8(qy78[mKate2::inx-8]) IV | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5131 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I;inx-8(qy78[mKate2::inx-8]) IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V; | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5133 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(qy102(mKate2::inx-8)) inx-9(ok1502) IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5136 | (this work) | sygl-1(q983[3xOLLAS::sygl-1]) naIs37[lag-2p::mCherry::PH +unc-119(+)] I; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5150 | (this work) | inx-14(ag17) sygl-1(q983[3xOLLAS::sygl-1]) naIs37[lag-2p::mCherry::PH +unc-119(+)] I; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5181 | (this work) | sygl-1(q983[3xOLLAS::sygl-1]) naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(qy78[mKate2::inx-8]) IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5229 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(qy78tn2031) IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5232 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(tn2034) IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5248 | (this work) | sygl-1(q983[3xOLLAS::sygl-1]) naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(qy78tn2031) IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5249 | (this work) | sygl-1(q983[3xOLLAS::sygl-1]) naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(tn2034) IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5250 | (this work) | inx-8(qy78tn2031) IV | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5251 | (this work) | inx-8(tn2034) IV | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5270 | (this work) | inx-14(ag17) I | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5310 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I; acy-4(ok1806) V; tnEx42[acy-4::gfp +rol-6(su1006)] | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5320 | (this work) | fasn-1(av138[fasn-1::gfp]) naIs37[lag-2p::mCherry::PH +unc-119(+)] I | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5346 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(qy78[mKate2::inx-8])/tmC5[tmIs1220] IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5347 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(qy78tn2031)/tmC5[tmIs1220] IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5357 | (this work) | tmC5[tmIs1220] inx-8(tn2075) IV | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5366 | (this work) | naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(qy78[mKate2::inx-8])/tmC5[tmIs1220] inx-8(tn2075) IV; bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5367 | (this work) | inx-14(ag17) fasn-1(av138[fasn-1::gfp]) naIs37[lag-2p::mCherry::PH +unc-119(+)] I | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5378 | (this work) | fasn-1(av138[fasn-1::gfp]) naIs37[lag-2p::mCherry::PH +unc-119(+)] I; inx-8(qy78[mKate2::inx-8]) IV | Contact DG for more information |
| Genetic reagent (C. elegans) | DG5380 | (this work) | bcIs39[lim-7p::ced-1::gfp +lin-15(+)] V | Contact DG for more information |
| Genetic reagent (C. elegans) | FX30140 | Dejima et al., 2018 | tmC5[tmIs1220] IV | Contact DG for more information |
| Genetic reagent (C. elegans) | JK1466 | Francis et al., 1995 | gld-1(q485)/dpy-5(e61) unc-32(e51) I | Contact DG for more information |
| Genetic reagent (C. elegans) | KLG006 | Gordon et al., 2020 | inx-8(qy78[mKate2::inx-8]) IV; tnIs6[plim-7::gfp +rol-6(su1006)] X; cpIs122(lag-2p::mNeonGreen::plcdeltaPH) | Contact DG for more information |
| Genetic reagent (C. elegans) | NK2571 | Gordon et al., 2020 | inx-8(qy78[mKate2::inx-8]); cpIs122 [lag-2p::mNeonGreen::plcdeltaPH] | Contact DG for more information |
| Genetic reagent (C. elegans) | NK2576 | Gordon et al., 2020 | inx-8(qy102(mKate2::inx-8)) inx-9(ok1502); cpIs122(lag-2p::mNeonGreen::plcdeltaPH) | Contact DG for more information |
Additional files
-
Supplementary file 1
Strains used in this study.
- https://cdn.elifesciences.org/articles/74955/elife-74955-supp1-v1.docx
-
Supplementary file 2
Oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/74955/elife-74955-supp2-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/74955/elife-74955-transrepform1-v1.docx