Evolutionary gain and loss of a plant pattern-recognition receptor for HAMP recognition
Figures
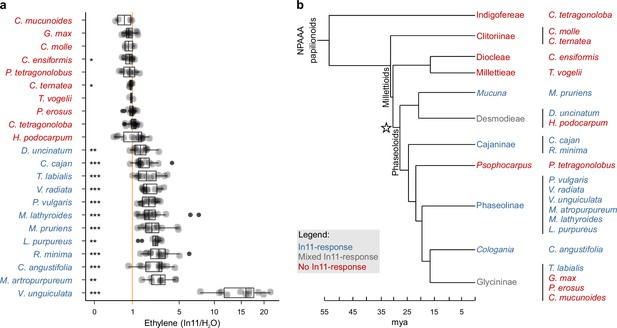
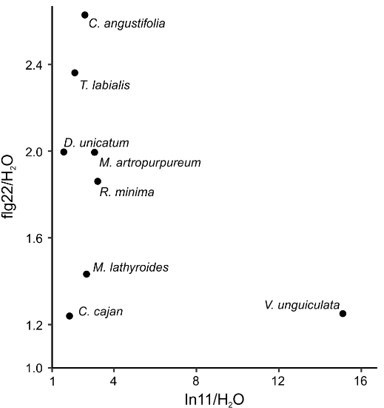
Induced ethylene response to In11 is limited to Phaseoloid legumes.
(a) Individual trifoliate leaflets were scratch wounded and treated with 1 μM In11 or H2O. The ratio of ethylene production for leaflets within the same leaf is shown (x-axis). The vertical orange line shows a ratio equal to one, i.e., no In11-induced ethylene burst. Biologically replicated plants are shown as separate dots. Significant differences between the control and the treatment of interest are indicated (paired Wilcoxon signed-rank test; ns non-significant, * p≤0.05, ** p≤0.01, and *** p≤0.001). Plant species names are colored blue (significant response to In11) or red (insignificant response). Species/accessions and resulting response data can respectively be found in Supplementary file 5a and Supplementary file 5b. (b) A summary chronogram representing time-based phylogenetic relationships within the tested lineages, with colors representing In11 response phenotypes as in (a). The star symbol indicates the node containing all In11-responsive species at the base of Phaseoloid legumes. Divergence time is shown in million years ago (mya) and represents a composite average (Egan et al., 2016; Li et al., 2013; Lavin et al., 2005; Stefanović et al., 2009).
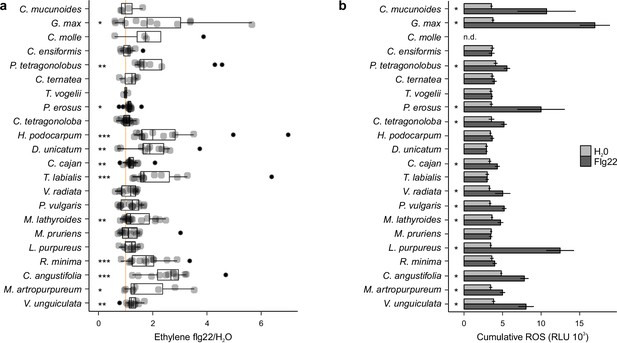
Flg22-induced reactive oxygen species (ROS) and ethylene responses are idiosyncratic across Millettioid and non-Millettioid legume species.
(a) Individual trifoliate leaflets were scratch wounded and treated with 1 μM flg22 or H2O. The ratio of ethylene production for leaflets within the same leaf is shown (x-axis). Species are listed in order of In11/H2O response as per Figure 1. The vertical orange line shows a ratio equal to one, i.e., no induced ethylene response to In11. Biological replicate plants are shown as separate dots. Significant differences between the control and the treatment of interest are indicated (paired Wilcoxon signed-rank test; ns non-significant, * p≤0.05, ** p≤0.01, and *** p≤0.001). (b) ROS-bursts are used a second marker of plant immunity response upon application of flg22. Shown is cumulative ROS data (3–60 min) of four technical replicates in relative luminescence units (RLUs) over 1 hr (1 observation/min) after treatment with H2O or the peptide flg22 (1 μM). No ROS data was determined for Centrosema molle (n.d.). Significant differences between the control and the treatment of interest were found by performing a two-group Mann-Whitney U Test (* p≤0.05).
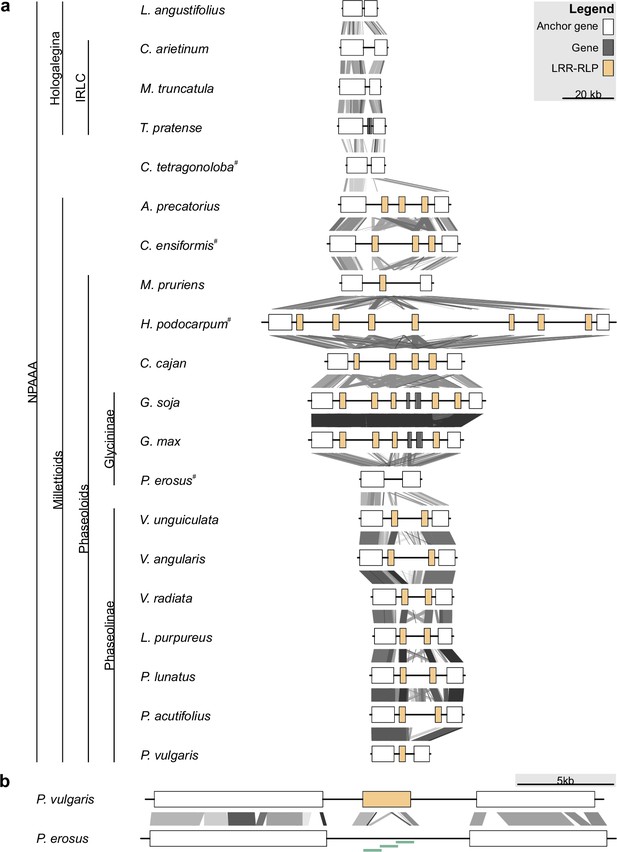
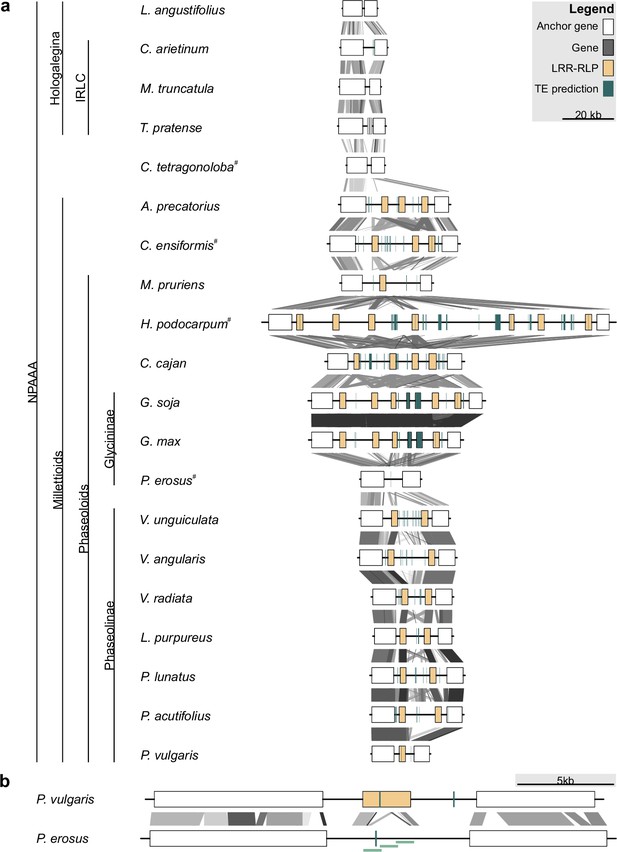
Leucine-rich repeat (LRR)-receptor-like protein (RLP) copy number variation at the INR locus in Millettioid and non-Millettioid legume genomes.
Anchor, LRR-RLP, and other genes are colored as per legend. (a) Locus comparison of the contiguous INR locus of 20 NPAAA papilionoid species. Blast hits between loci are indicated with lines (e-value <1e-04) with score according to grayscale gradient and darker grays indicating higher similarity. Genes are labeled LRR-RLP if a complete coding sequence is present (≥875 AA). Species names followed by superscript (#) were newly sequenced and assembled for this project. (b) Locus comparison of the contiguous INR locus of P. vulgaris and P. erosus. P. vulgaris has a functionally validated inceptin receptor (INR) homolog (LRR-RLP) (Steinbrenner et al., 2020), while P. erosus lacks a full-length LRR-RLP. The disruption of the P. erosus LRR-RLP was validated by PCR followed by Sanger sequencing as indicated with light green bars.
Repetitive elements at the INR locus in Millettioid and non-Millettioid legume genomes.
CENSOR was used to perform a rapid identification of repetitive elements in between the anchor genes of the depicted INR loci by comparison to known repeats of the Arabidopsis thaliana Repbase database (Kohany et al., 2006). Detailed output of the analysis can be found in the Supplementary file 6c.
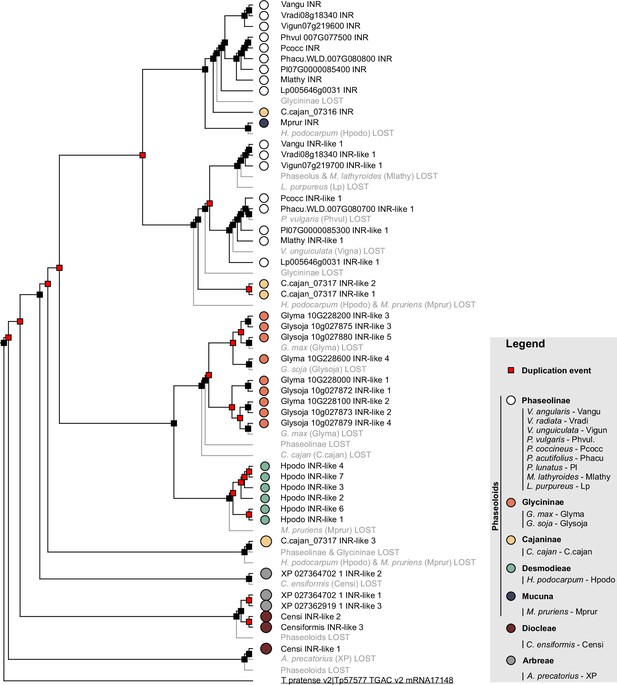
Prediction of gene duplication and gene loss events throughout the evolution of the contiguous INR locus in the Millettioids.
Gene tree-species tree reconciliation to identify the duplication and loss events at each branch using NOTUNG (Chen et al., 2000). Predicted duplication events are marked with a red square, black squares represent speciation events, and lost nodes/genes are highlighted in gray. Filled circles indicate species of origin according to legend, where different colors indicate different subtribes. A T. pratense leucine-rich repeat (LRR)-receptor-like protein (RLP) was used as an outgroup to root the phylogenetic gene tree and is underlined.
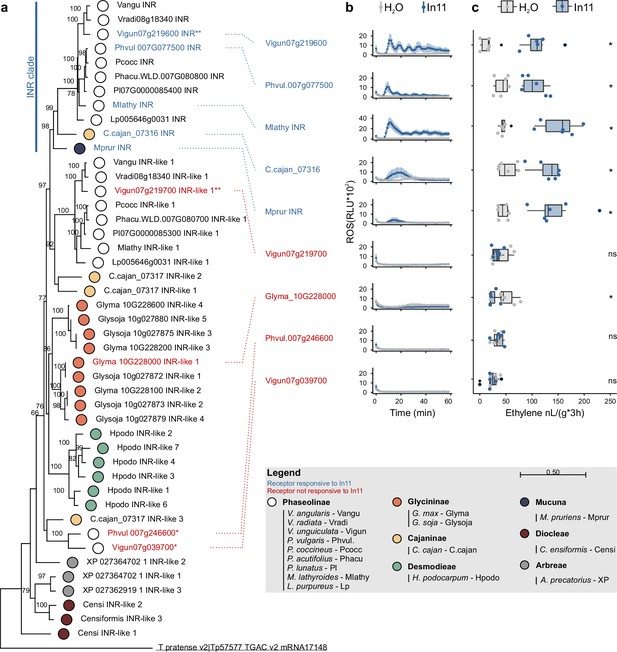
Phylogenetic analysis of leucine-rich repeat (LRR)-receptor-like proteins (RLPs) at the contiguous INR locus in 16 diverse species and subsequent heterologous expression reveals a clade of functional receptors.
(a) A phylogenetic analysis of LRR-RLPs from 16 Millettioid species is shown. Maximum likelihood analysis bootstrap values are indicated, and only values higher than 65 are shown. The scale bar represents 0.5 AA substitutions per site. Filled dots indicate species of origin according to legend, where different colors indicate different subtribes. A T. pratense LRR-RLP was used as an outgroup to root the phylogenetic gene tree and is underlined. One asterisk (*) highlights the LRR-RLPs which are not part of a contiguous INR locus. Two asterisks (**) highlight the LRR-RLPs used to create the chimeric receptors. The functionally validated inceptin receptors (INRs) of P. vulgaris, V. unguiculata, M. lathyroides, C. cajan, and M. pruriens are highlighted in blue as they confer induced reactive oxygen species (ROS) and ethylene functions in response to In11 upon heterologous expression in N. benthamiana, as shown respectively in panel B and C. These five validated INRs fall within the labeled ‘INR clade’. Heterologously expressed receptors which were not responsive to In11 are highlighted in red. (b) Shown are relative luminescence units (RLUs) after treatment with H2O (gray) or the peptide In11 (1 μM, blue). Curves indicate mean ± SD for four independent biological replicates (n=4 plants), with each biological replicate representing six technical replicates. (c) The x-axis shows the amount of ethylene released after infiltration with 1 μM In11 (blue) or water (gray). Dots represent independent biological replicates (n=6 plants). Significance was tested by performing a paired Wilcoxon signed-rank test (ns non-significant, * p≤0.05).

Western blot of the heterologously expressed constructs in N. benthamiana.
Tissue was harvested 48 hr after construct infiltration in N. benthamiana. The western blot was probed with (1) GFP antibody as the receptor constructs had a C-terminal GFP tag (top) and (2) actin antibody as a loading control (bottom). The western blot shows all heterologously expressed receptors inceptin receptor (INR) and INR-like homologs from diverse legumes of which response to In11 was tested. See Figure 3—figure supplement 1—source data 1 for original files.
-
Figure 3—figure supplement 1—source data 1
Original and labeled western blot of the heterologously expressed inceptin receptor (INR) and INR-like homolog constructs in N. benthamiana.
For the western blot of C-terminal GFP tagged INR and INR-like homolog receptors, after wet transfer, nitrocellulose membranes were physically split at 75 kDA and probed separately with (1) GFP antibody (>75 kDa) and (2) actin antibody as a loading control (<75 kDa), resulting in two tiff files for each panel.
- https://cdn.elifesciences.org/articles/81050/elife-81050-fig3-figsupp1-data1-v2.zip
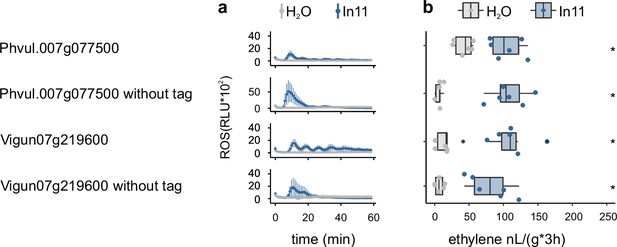
The effect of a C-terminal GFP tag on In11-dependent reactive oxygen species (ROS) and ethylene burst of inceptin receptor (INR).
(a) In11-dependent ROS production following the heterologous expression of receptors in N. benthamiana. Shown are relative luminescence units (RLUs) after treatment with H2O (gray) or the peptide In11 (1 μM, blue). Curves indicate mean ± SD for four independent biological replicates (n=4 plants), with each biological replicate representing six technical replicates. (b) Ethylene production following the heterologous expression of receptors in N. benthamiana. Ethylene production was quantified after infiltration with H2O (gray) or the peptide In11 (1 μM, blue). Dots represent independent biological replicates (n=6 plants). Significance was tested by performing a paired Wilcoxon signed-rank test (ns non-significant, * p≤0.05).
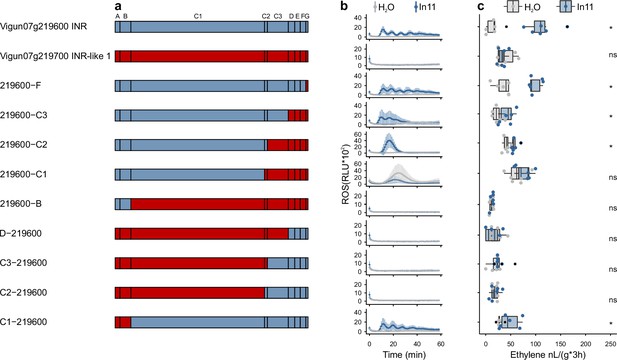
Chimeric receptors indicate that the C1 and C2 subdomains mediate inceptin receptor (INR) recognition function.
(a) Schematic representation of Vigun07g219600 (blue, VuINR), Vigun07g219700 (red, VuINR-like), and the nine created chimeric receptors used for structure-function analysis. Leucine-rich repeat (LRR)-receptor-like protein (RLP) subdomains as per Fritz-Laylin et al., 2005; A: putative signal peptide, B: one or two pairs of Cys that may play structural roles, C: multiple LRRs with an intervening motif (C2) inserted, D: linker domain, E: acidic domain, F: transmembrane helix, and G: cytoplasmic tail. Nucleotide sequences of the chimeric receptors can be found in Supplementary file 3. (b) In11-dependent reactive oxygen species (ROS) production following the heterologous expression of receptors in N. benthamiana. Shown are relative luminescence units (RLUs) after treatment with H2O (gray) or the peptide In11 (1 μM, blue). Curves indicate mean ± SD for four independent biological replicates (n=4 plants), with each biological replicate representing six technical replicates. (c) Ethylene production following the heterologous expression of receptors in N. benthamiana. Ethylene production was quantified after infiltration with H2O (gray) or the peptide In11 (1 μM, blue). Dots represent independent biological replicates (n=6 plants). Significance was tested by performing a paired Wilcoxon signed-rank test (ns non-significant, * p≤0.05).
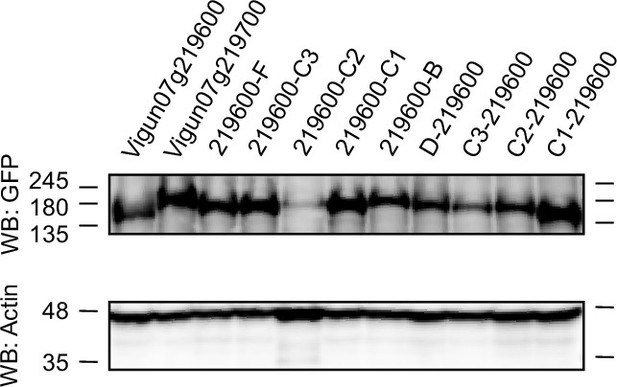
Western blot of the heterologously expressed chimeric receptor constructs in N. benthamiana.
Tissue was harvested 48 hr after construct infiltration in N. benthamiana. The western blot was probed with (1) GFP antibody as the receptor constructs had a C-terminal GFP tag (top) and (2) actin antibody as a loading control (bottom). The western blot shows all heterologously expressed chimeric receptors of which response to In11 was tested: See Figure 4—figure supplement 1—source data 1 for original files.
-
Figure 4—figure supplement 1—source data 1
Original and labeled western blot of the heterologously expressed chimeric receptor constructs in N. benthamiana.
For the western blot of C-terminal GFP tagged chimeric receptors, after wet transfer, nitrocellulose membranes were physically split at 75 kDA and probed separately with (1) GFP antibody (>75 kDa) and (2) actin antibody as a loading control (<75 kDa), resulting in two tiff files for each panel.
- https://cdn.elifesciences.org/articles/81050/elife-81050-fig4-figsupp1-data1-v2.zip
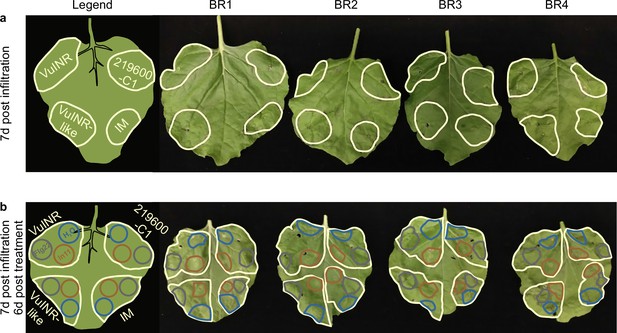
Heterologous expression of chimeric receptor 219600-C1 does not result in a visible phenotype in N. benthamiana.
Pictures were taken 7 days post-infiltration of Agrobacterium strains encoding the chimeric receptor 219600-C1, its parental receptors VuINR (Vigun07g219600), and VuINR-like1 (Vigun07g219700), or solely IM: infiltration media. (a) No subsequent treatments were performed after initial construct infiltration. (b) 1 day after Agrobacterium infiltration, the following treatments were infiltrated: 1 µM flg22 (gray), 1 µM In11 (brown), or H2O (blue).
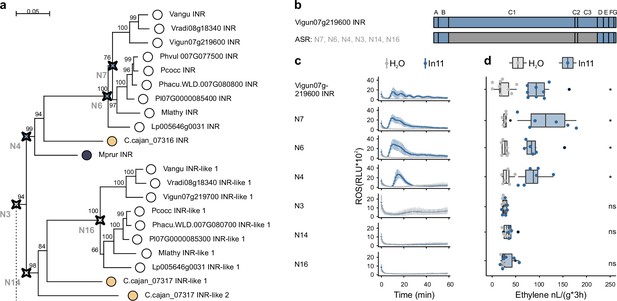
Functional analysis of ancestrally reconstructed leucine-rich repeat (LRR) domains of inceptin receptor (INR) and INR-like receptors.
(a) Part of the phylogenetic analyses of the LRR (C1-3 domain) of LRR-receptor-like proteins (RLPs) from the contiguous INR locus used for the ancestral sequence reconstruction (ASR) full phylogeny can be found in Figure 5—figure supplement 1. Ancestrally reconstructed nodes are marked with a compass star (N7, N6, N4, N3, N14, and N16). The scale bar represents 0.05 AA substitutions per site. Nucleotide sequences of the LRR domain can be found in Supplementary file 4. (b) Schematic representation of Vigun07g219600 (blue, VuINR) and the six created ASR receptors used for structure-function analysis. LRR-RLP subdomains as per Fritz-Laylin et al., 2005; A: putative signal peptide, B: one or two pairs of Cys that may play structural roles, C: multiple LRRs with an intervening motif (C2) inserted, D: linker domain, E: acidic domain, F: transmembrane helix, and G: cytoplasmic tail. Nucleotide sequences of the chimeric receptors can be found in Supplementary file 3. (c) In11-dependent reactive oxygen species (ROS) production following the heterologous expression of the ASR receptors in N. benthamiana. Relative luminescence units (RLUs) are shown after treatment with H2O (gray), or the peptide In11 (1 μM, blue). Curves indicate mean ± SD for four independent biological replicates (n=4 plants), with each biological replicate representing six technical replicates. (d) Ethylene production following the heterologous expression of receptors in N. benthamiana. Ethylene production was quantified after infiltration with H2O (gray) or the peptide In11 (1 μM, blue). Dots represent independent biological replicates (n≥6 plants). Significance was tested by performing a paired Wilcoxon signed-rank test (ns non-significant, * p≤0.05).
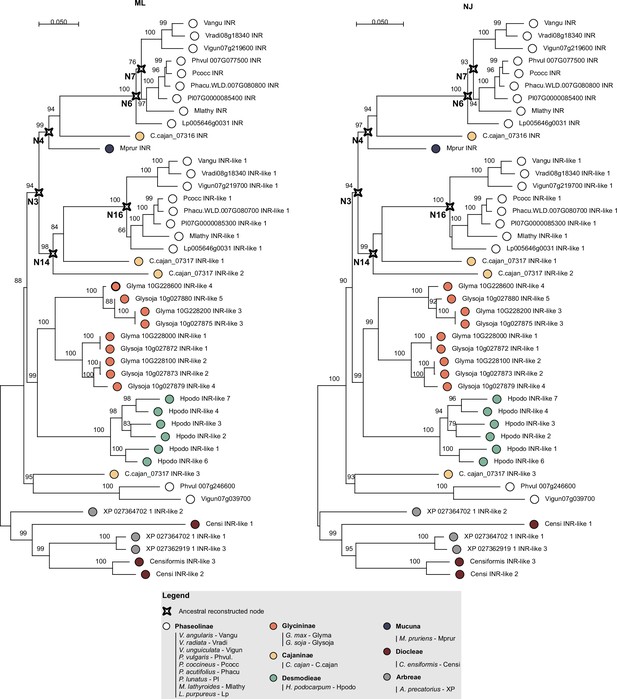
Phylogenetic analyses of the leucine-rich repeat (LRR) (C1-3 domain) of LRR-receptor-like proteins (RLPs) from the contiguous INR locus.
The phylogenetic trees were built using MEGA X software and 1000 bootstraps (Kumar et al., 2018). Maximum likelihood (ML – left side) and neighbor joining (NJ – right side) trees were calculated based on all codon positions of a codon-based alignment (Edgar, 2004). ML analysis bootstrap values are indicated; only values higher than 65 are shown. The scale bar represents 0.05 AA substitutions per site. The ancestral reconstructed nodes selected for functional validation are marked with a compass star (N7, N6, N4, N3, N14, and N16). Filled dots indicate species of origin according to legend, where different colors indicate different subtribes.
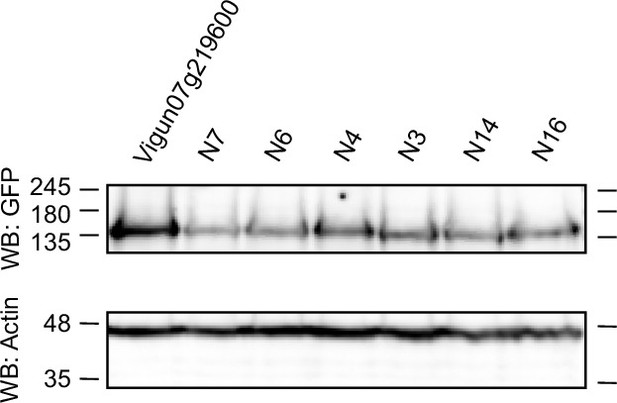
Western blot of the heterologously expressed ancestral sequence reconstruction (ASR) receptor constructs in N. benthamiana.
Tissue was harvested 48 hr after construct infiltration in N. benthamiana. The western blot was probed with (1) GFP antibody as the receptor constructs had a C-terminal GFP tag (top) and (2) actin antibody as a loading control (bottom). The western blot shows all heterologously expressed ASR receptors of which response to In11 was tested. See Figure 5—figure supplement 2—source data 1 for original files.
-
Figure 5—figure supplement 2—source data 1
Original and labeled western blot of the heterologously expressed ancestral sequence reconstruction (ASR) receptor constructs in N. benthamiana.
For the western blot of C-terminal GFP tagged ASR receptors, after wet transfer, nitrocellulose membranes were physically split at 75 kDA and probed separately with (1) GFP antibody (>75 kDa) and (2) actin antibody as a loading control (<75 kDa), resulting in two tiff files for each panel.
- https://cdn.elifesciences.org/articles/81050/elife-81050-fig5-figsupp2-data1-v2.zip
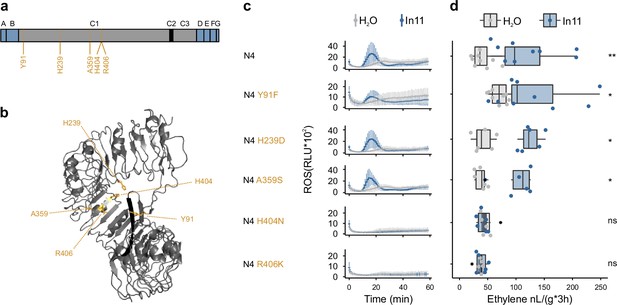
The H404N and R406K AA substitutions abolish the function of N4, the ancestor of all inceptin receptor (INR)-homologs.
(a) Schematic representation of the N4 ancestral sequence reconstruction (ASR) receptor used for structure-function analysis. Five strongly conserved amino acid (AA) in all INR-homologs and N4 which differ from the corresponding N3 AA are highlighted in yellow. Leucine-rich repeat (LRR)-receptor-like protein (RLP) subdomains as per Fritz-Laylin et al., 2005; A: putative signal peptide, B: one or two pairs of Cys that may play structural roles, C1: multiple LRRs (colored in gray) with an intervening motif (C2, colored in black) inserted, D: linker domain, E: acidic domain, F: transmembrane helix, and G: cytoplasmic tail. (b) AlphaFold prediction of VuINR (Vigun07g219600), only the C-domain is depicted with the C2-domain colored in black. Similar as in (a), the five strongly conserved AAs in all INR-homologs and N4 which differ from the corresponding N3 AAs are highlighted in yellow. (c) In11-dependent reactive oxygen species (ROS) production following the heterologous expression of the N4 and N4 variant receptors in N. benthamiana. Relative luminescence units (RLUs) are shown after treatment with H2O (gray) or the peptide In11 (1 μM, blue). Curves indicate mean ± SD for four independent biological replicates (n=4 plants), with each biological replicate representing six technical replicates. (d) Ethylene production following the heterologous expression of receptors in N. benthamiana. Ethylene production was quantified after infiltration with H2O (gray) or the peptide In11 (1 μM, blue). Dots represent independent biological replicates (n≥6 plants). Significance was tested by performing a paired Wilcoxon signed-rank test (ns non-significant, * p≤0.05).
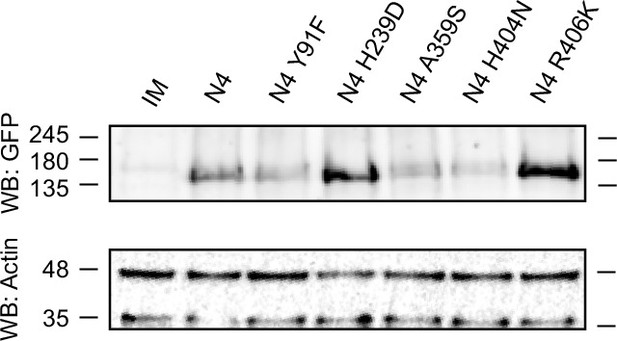
Western blot of the heterologously expressed N4 and N4 variant constructs in N. benthamiana.
Tissue was harvested 48 hr after construct infiltration in N. benthamiana. The western blot was probed with (1) GFP antibody as the receptor constructs had a C-terminal GFP tag (top) and (2) actin antibody (A0480) as a loading control (bottom). The western blot shows all heterologously expressed N4 receptors with/without single amino acid (AA) substitutions of which response to In11 was tested and an infiltration media only control (IM). See Figure 6—figure supplement 1—source data 1 for original files.
-
Figure 6—figure supplement 1—source data 1
Original and labeled western blot of the heterologously expressed N4 and N4 variant constructs in N. benthamiana.
Two identical western blots were run in parallel of the C-terminal GFP tagged N4 and N4 variant constructs receptors and probed differentially with (1) GFP antibody and (2) actin antibody (A0480) as a loading control, resulting in two tiff files for each blot.
- https://cdn.elifesciences.org/articles/81050/elife-81050-fig6-figsupp1-data1-v2.zip
Predicted LDDT(Local distance difference test) score per position for all five created AlphaFold models of Vigun07g219600.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81050/elife-81050-mdarchecklist1-v2.docx
-
Supplementary file 1
Fasta file of nt sequences of the inceptin receptor (INR) syntenic loci incorporated in the contiguous INR locus analysis.
- https://cdn.elifesciences.org/articles/81050/elife-81050-supp1-v2.zip
-
Supplementary file 2
Fasta file of the amino acid (AA) sequences inceptin receptor (INR) and INR-like homologs included in the phylogenetic analysis.
- https://cdn.elifesciences.org/articles/81050/elife-81050-supp2-v2.zip
-
Supplementary file 3
Fasta file containing the sequences of the chimeric receptors.
- https://cdn.elifesciences.org/articles/81050/elife-81050-supp3-v2.zip
-
Supplementary file 4
Fasta file containing the nucleotide leucine-rich repeat (LRR) domain sequences of the inceptin receptor (INR) and INR-like homologs used for the ancestral sequence reconstruction (ASR) analysis and the resulting predicted (and domesticated for MoClo) LRR domain sequences for the ancestral nodes of interest.
- https://cdn.elifesciences.org/articles/81050/elife-81050-supp4-v2.zip
-
Supplementary file 5
Excel file containing (A) overview of the used legume species, accessions, and their origin, and (B) overview of ethylene response data of legume species.
- https://cdn.elifesciences.org/articles/81050/elife-81050-supp5-v2.xlsx
-
Supplementary file 6
Excel file containing (A) overview of mined assemblies of contiguous inceptin receptor (INR) loci (+ coordinates) and leucine-rich repeat (LRR) receptor-like proteins (RLPs), (B) genome assembly stats, and (C) output file of CENSOR analysis which detected repetitive elements of the INR loci by comparison to known repeats of the Arabidopsis thaliana Repbase database.
- https://cdn.elifesciences.org/articles/81050/elife-81050-supp6-v2.xlsx
-
Supplementary file 7
Excel file containing (A) primers used in this study, and (B) p-values of statistical analysis depicted in all figures and figure supplements.
- https://cdn.elifesciences.org/articles/81050/elife-81050-supp7-v2.xlsx
-
Supplementary file 8
Amino acid (AA) sequence alignment of the leucine-rich repeat (LRR) domains of the inceptin receptor (INR) clade, INR-like 1, clade and the closest ancestral reconstructed nodes with differential In11-response.
Differences or consensus (dashed marks) from ancestral sequence reconstruction (ASR) construct N3 are indicated for all other sequences. The five AA polymorphisms between N3 and N4 that are similar within all LRR-RLPs of the INR clade and different in LRR-RLPs of the INR-like 1 sister clade are highlighted in yellow. C1-domain starts at the N-terminus, C3-domain ends at the C-terminus, C1 and C3 domain are separated by the C2-domain which is highlighted in the frame.
- https://cdn.elifesciences.org/articles/81050/elife-81050-supp8-v2.pdf