Effector target-guided engineering of an integrated domain expands the disease resistance profile of a rice NLR immune receptor
Figures
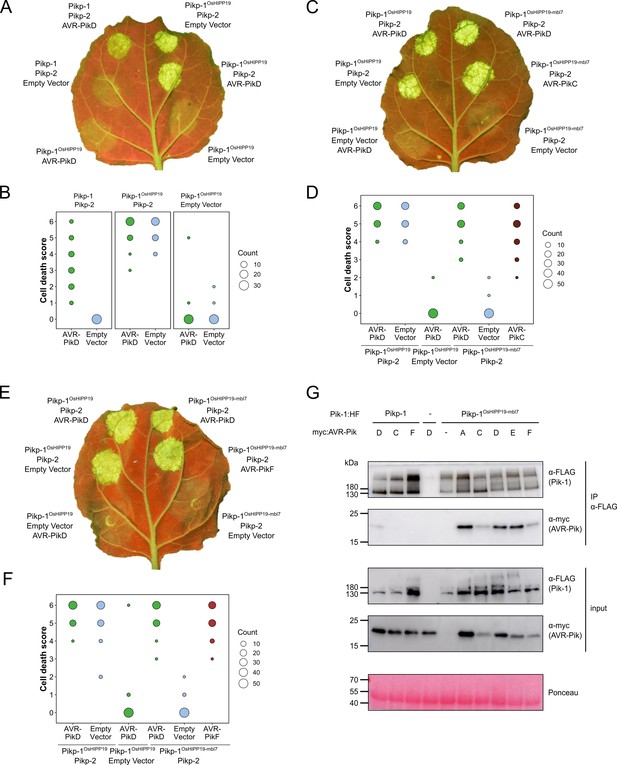
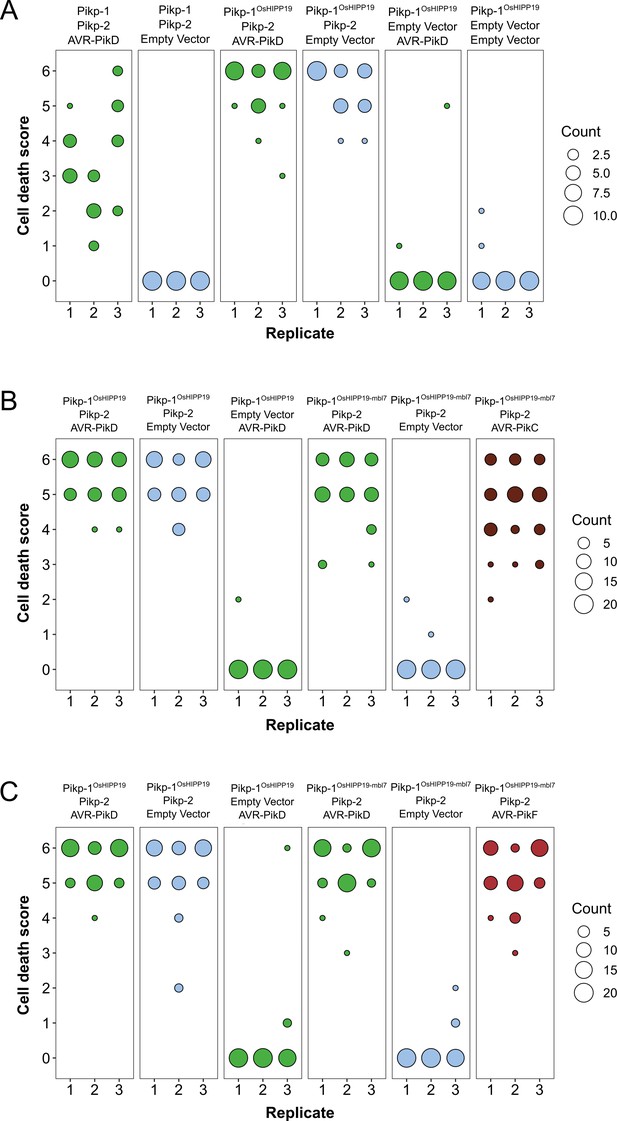
The Pikp-1OsHIPP19-mbl7 chimera expands binding and response to previously unrecognized AVR-Pik effector variants.
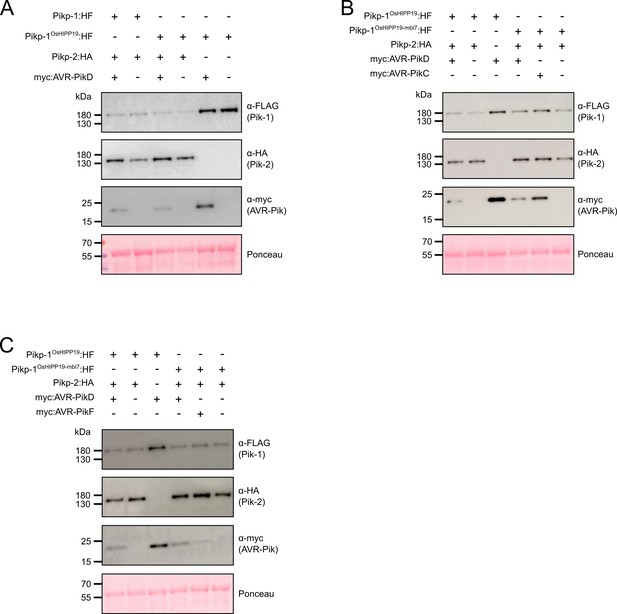
(A) Representative leaf image showing the Pikp-1OsHIPP19 chimera is autoactive in N. benthamiana in a Pikp-2 dependent manner. Nucleotide-binding and leucine-rich repeat (NLR)-mediated responses appear as autofluorescence imaged under UV light. Pikp-mediated response to AVR-PikD (positive control, top left), Pikp-1OsHIPP19/Pikp-2 without effector shows autoactivity (top right), Pikp-1OsHIPP19/Pikp-2 response remains in the presence of AVR-PikD (middle right). Other leaf positions represent relevant negative controls. (B) Pikp-mediated response scoring represented as dot plots to summarize 30 repeats of the experiment shown in (A) across three independent experiments (Materials and methods, Figure 1—figure supplement 4a). Fluorescence intensity is scored as previously described (Maqbool et al., 2015; De la Concepcion et al., 2018). (C) The Pikp-1OsHIPP19-mbl7 chimera does not display autoactive cell death in N. benthamiana (bottom right), as seen for Pikp-1OsHIPP19 (middle left), but retains response to AVR-PikD and expands Pikp-mediated response to AVR-PikC (middle right). (D) Pikp-mediated response scoring represented as dot plots to summarize 60 repeats of the experiment shown in (C) across three independent experiments (Materials and methods, Figure 1—figure supplement 4b). (E) As for (C), but showing the expanded Pikp-mediated response to AVR-PikF. (F) As for (D) but for 60 repeats of the experiment in (E) across three independent experiments (Materials and methods, Figure 1—figure supplement 4c). (G) Western blots following co-immunoprecipitation reveal that the Pikp-1OsHIPP19-mbl7 chimera associates with all AVR-Pik effector variants in N. benthamiana. Plant cell lysates were probed for the expression of Pikp-1/Pikp-1OsHIPP19-mbl7 and AVR-Pik effector variants using anti-FLAG and anti-Myc antiserum, respectively. Total protein extracts were visualized by Ponceau Staining.
-
Figure 1—source data 1
Unedited and uncropped blot for panel G, IP Pik-1 α-FLAG, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data1-v1.tiff
-
Figure 1—source data 2
Unedited and uncropped blot for panel G, IP Pik-1 α-FLAG, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data2-v1.tiff
-
Figure 1—source data 3
Unedited and uncropped blot for panel G, IP Pik-1 α-FLAG, AVRPik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data3-v1.tiff
-
Figure 1—source data 4
Unedited and uncropped blot for panel G, IP Pik-1 α-FLAG, AVRPik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data4-v1.tiff
-
Figure 1—source data 5
Unedited and uncropped blot for panel G, input, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data5-v1.tiff
-
Figure 1—source data 6
Unedited and uncropped blot for panel G, input, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data6-v1.tiff
-
Figure 1—source data 7
Unedited and uncropped blot for panel G, input, AVRPik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data7-v1.tiff
-
Figure 1—source data 8
Unedited and uncropped blot for panel G, input, AVRPik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data8-v1.tiff
-
Figure 1—source data 9
Unedited and uncropped blot for panel G, Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data9-v1.tiff
-
Figure 1—source data 10
Unedited and uncropped blot for panel G, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data10-v1.tiff
-
Figure 1—source data 11
Cell death scores used for dot plots in panel B.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data11-v1.zip
-
Figure 1—source data 12
Cell death scores used for dot plots in panel D.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data12-v1.zip
-
Figure 1—source data 13
Cell death scores used for dot plots in panel F.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-data13-v1.zip
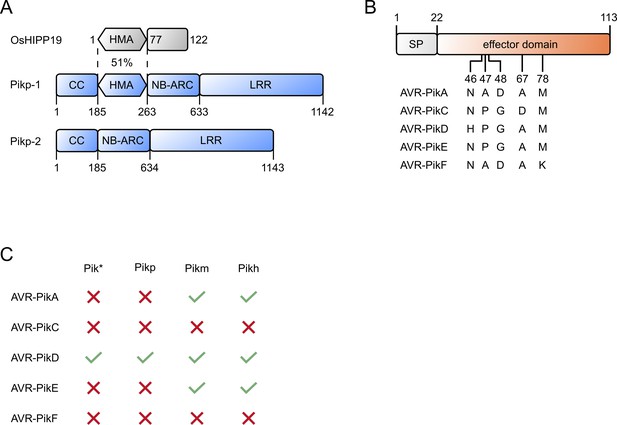
Domain structure and resistance profiles for the proteins in this study.
(A) Cartoon representation of the OsHIPP19, Pik-1, and Pik-2 domains, with numbers giving the amino acid domain boundaries. CC = Coiled coil, HMA = Heavy-metal-associated, NB-ARC = Nucleotide-binding found in APAF-1, R proteins and CED4, LRR = Leucine rich repeat. (B) Cartoon representation of AVR-Pik effector variants. Amino acid polymorphisms between variants are shown as single-letter codes with the number giving the position. SP = signal peptide. (C) Summary of the recognition profiles of known Pik nucleotide-binding and leucine-rich repeat (NLR) alleles against AVR-Pik variants (tick = resistant, cross = susceptible).
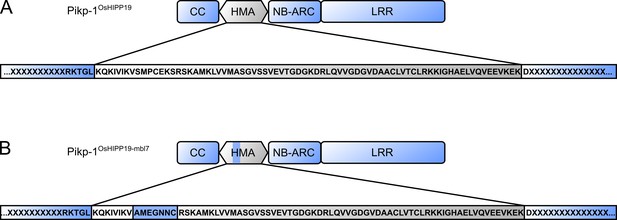
Schematic representation of the Pikp-1OsHIPP19 (A) and Pikp-1OsHIPP19-mbl7 (B) chimeras.
The amino acid sequence below indicates junctions between the sequence derived from Pikp-1 (blue highlight) and from OsHIPP19 (gray highlight).
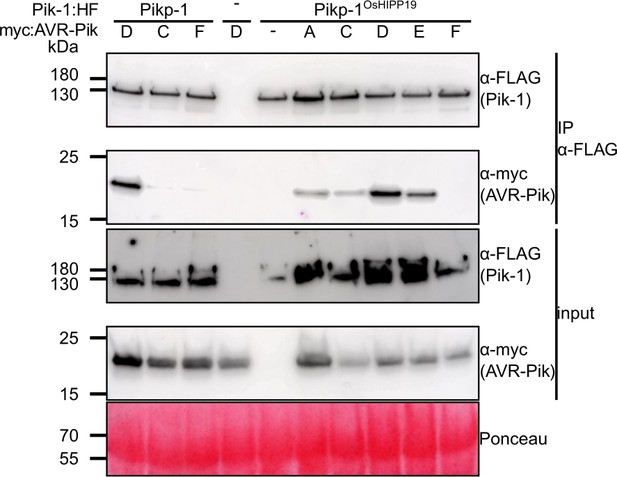
Western blots following co-immunoprecipitation show that the Pikp-1OsHIPP19 chimera binds to all AVR-Pik effector variants except, surprisingly, AVR-PikF in N. benthamiana.
Plant cell lysates were probed for the expression of Pikp-1/Pikp-1OsHIPP19 and AVR-Pik effector variants using anti-FLAG and anti-Myc antiserum, respectively. Total protein extracts were visualized by Ponceau Staining.
-
Figure 1—figure supplement 3—source data 1
Unedited and uncropped blot, IP Pik-1 α-FLAG, Pik-1 α-FLAG, with relevant bands labelled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp3-data1-v1.tiff
-
Figure 1—figure supplement 3—source data 2
Unedited and uncropped blot, IP Pik-1 α-FLAG, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp3-data2-v1.tiff
-
Figure 1—figure supplement 3—source data 3
Unedited and uncropped blot, IP Pik-1 α-FLAG, AVRPik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp3-data3-v1.tiff
-
Figure 1—figure supplement 3—source data 4
Unedited and uncropped blot, IP Pik-1 α-FLAG, AVRPik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp3-data4-v1.tiff
-
Figure 1—figure supplement 3—source data 5
Unedited and uncropped blot, input, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp3-data5-v1.tiff
-
Figure 1—figure supplement 3—source data 6
Unedited and uncropped blot, input, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp3-data6-v1.tiff
-
Figure 1—figure supplement 3—source data 7
Unedited and uncropped blot, input, AVRPik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp3-data7-v1.tiff
-
Figure 1—figure supplement 3—source data 8
Unedited and uncropped blot, input, AVRPik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp3-data8-v1.tiff
-
Figure 1—figure supplement 3—source data 9
Unedited and uncropped blot, Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp3-data9-v1.tiff
-
Figure 1—figure supplement 3—source data 10
Unedited and uncropped blot, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp3-data10-v1.tiff
Pikp-mediated response scoring is represented as dot plots, subdivided by replicate, for repeats of experiments presented in Figure 1a, c and e (panels A, B, and C; respectively).
Each replicate consisted of 10 (A) or 20 (B, C) repeats for each sample. Fluorescence intensity is scored as described in Figure 1. Scores from the three replicates in panels A, B, and C were combined and represented as the dot plots in Figure 1b, d and f, respectively.
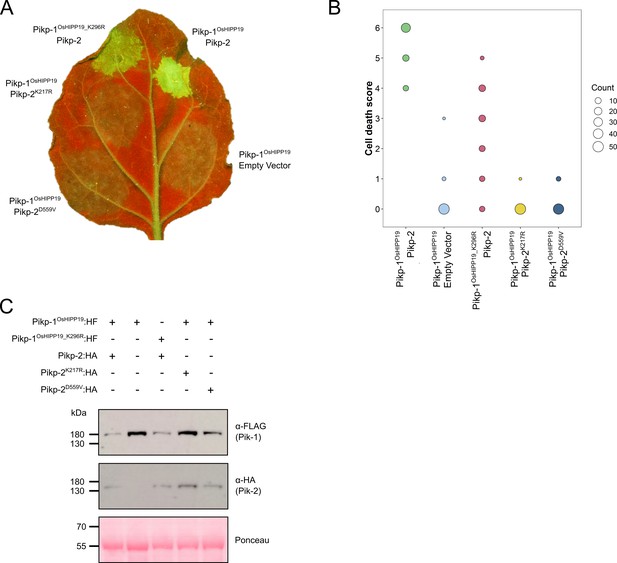
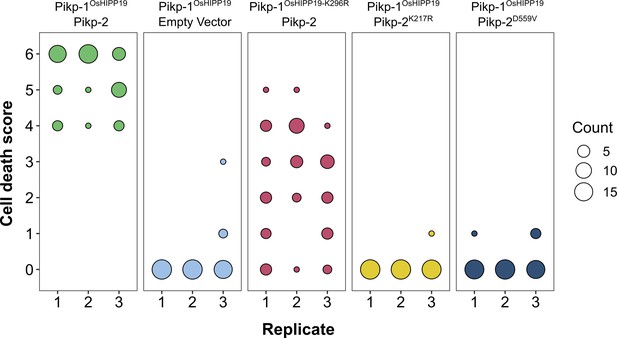
The Pikp-1OsHIPP19 chimera requires both the P-loop and MHD motifs in Pikp-2 for autoactivity, and the P-loop in Pikp-1 for full cell death.
(A) Example leaf showing the P-loop mutant (K217R, middle, left) and MHD mutant (D559V, bottom, left) in Pikp-2 abolishes autoactive cell death, whereas the P-loop mutant in Pikp-1OsHIPP19 (K196R, top, left) reduces the cell death response. (B) Pikp-mediated response scoring represented as dot plots to summarize 54 repeats of the experiment shown in (A) across three independent experiments (Materials and methods, Figure 1—figure supplement 6). Fluorescence intensity is scored as stated in Figure 1. (C) Western blots confirming the accumulation of proteins in N. benthamiana. Plant cell lysates were probed for the expression of Pikp-1, Pikp-2, and mutants thereof, using anti-FLAG, and anti-HA antiserum, respectively. Total protein extracts were visualized by Ponceau Staining.
-
Figure 1—figure supplement 5—source data 1
Unedited and uncropped blot for panel C, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp5-data1-v1.tiff
-
Figure 1—figure supplement 5—source data 2
Unedited and uncropped blot for panel C, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp5-data2-v1.tiff
-
Figure 1—figure supplement 5—source data 3
Unedited and uncropped blot for panel C, Pik-2 α-HA, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp5-data3-v1.tiff
-
Figure 1—figure supplement 5—source data 4
Unedited and uncropped blot for panel C, Pik-2 α-HA.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp5-data4-v1.tiff
-
Figure 1—figure supplement 5—source data 5
Unedited and uncropped blot for panel C, Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp5-data5-v1.tiff
-
Figure 1—figure supplement 5—source data 6
Unedited and uncropped blot for panel C, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp5-data6-v1.tiff
-
Figure 1—figure supplement 5—source data 7
Cell death scores used for dot plots for panel B.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp5-data7-v1.zip
Pikp-mediated response scoring is represented as dot plots, subdivided by replicate, for repeats of the experiment presented in Figure 1—figure supplement 5a.
Each replicate consisted of 18 repeats for each sample. Fluorescence intensity is scored as described in Figure 1. Scores from the three replicates were combined and represented as the dot plot in Figure 1—figure supplement 5b.
Western blots confirm the accumulation of proteins in N. benthamiana for the cell death assays shown in Figure 1.
Plant cell lysates were probed for the expression of Pikp-1/Pikp-1OsHIPP19/Pikp-1OsHIPP19-mbl7, Pikp-2, and AVR-Pik effector variants using anti-FLAG, anti-HA, and anti-Myc antiserum, respectively. Total protein extracts were visualized by Ponceau Staining.
-
Figure 1—figure supplement 7—source data 1
Unedited and uncropped blot for panel A, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data1-v1.tiff
-
Figure 1—figure supplement 7—source data 2
Unedited and uncropped blot for panel A, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data2-v1.tiff
-
Figure 1—figure supplement 7—source data 3
Unedited and uncropped blot for panel A, Pik-2 α-HA, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data3-v1.tiff
-
Figure 1—figure supplement 7—source data 4
Unedited and uncropped blot for panel A, Pik-2 α-HA.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data4-v1.tiff
-
Figure 1—figure supplement 7—source data 5
Unedited and uncropped blot for panel A, AVR-Pik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data5-v1.tiff
-
Figure 1—figure supplement 7—source data 6
Unedited and uncropped blot for panel A, AVR-Pik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data6-v1.tiff
-
Figure 1—figure supplement 7—source data 7
Unedited and uncropped blot for panel A, Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data7-v1.tiff
-
Figure 1—figure supplement 7—source data 8
Unedited and uncropped blot for panel A, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data8-v1.tiff
-
Figure 1—figure supplement 7—source data 9
Unedited and uncropped blot for panel B, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data9-v1.tiff
-
Figure 1—figure supplement 7—source data 10
Unedited and uncropped blot for panel B, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data10-v1.tiff
-
Figure 1—figure supplement 7—source data 11
Unedited and uncropped blot for panel B, Pik-2 α-HA, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data11-v1.tiff
-
Figure 1—figure supplement 7—source data 12
Unedited and uncropped blot for panel B, Pik-2 α-HA.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data12-v1.tiff
-
Figure 1—figure supplement 7—source data 13
Unedited and uncropped blot for panel B, AVR-Pik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data13-v1.tiff
-
Figure 1—figure supplement 7—source data 14
Unedited and uncropped blot for panel B, AVR-Pik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data14-v1.tiff
-
Figure 1—figure supplement 7—source data 15
Unedited and uncropped blot for panel B, Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data15-v1.tiff
-
Figure 1—figure supplement 7—source data 16
Unedited and uncropped blot for panel B, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data16-v1.tiff
-
Figure 1—figure supplement 7—source data 17
Unedited and uncropped blot for panel C, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data17-v1.tiff
-
Figure 1—figure supplement 7—source data 18
Unedited and uncropped blot for panel C, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data18-v1.tiff
-
Figure 1—figure supplement 7—source data 19
Unedited and uncropped blot for panel C, Pik-2 α-HA, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data19-v1.tiff
-
Figure 1—figure supplement 7—source data 20
Unedited and uncropped blot for panel C, Pik-2 α-HA.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data20-v1.tiff
-
Figure 1—figure supplement 7—source data 21
Unedited and uncropped blot for panel C, AVR-Pik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data21-v1.tiff
-
Figure 1—figure supplement 7—source data 22
Unedited and uncropped blot for panel C, AVR-Pik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data22-v1.tiff
-
Figure 1—figure supplement 7—source data 23
Unedited and uncropped blot for panel C.
Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data23-v1.tiff
-
Figure 1—figure supplement 7—source data 24
Unedited and uncropped blot for panel C, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp7-data24-v1.tiff
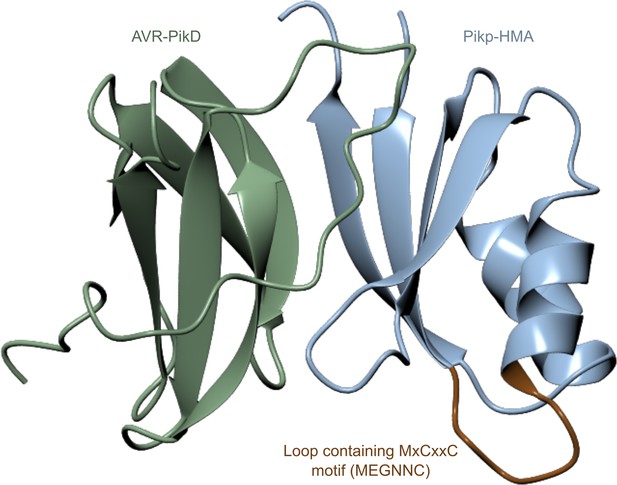
Location of the β1-α1 loop (brown) in Pikp-HMA (blue) is distant from the effector (green) binding surface in the crystal structure of complexes between these proteins.
Structure shown is based on PDB entry 6G10. Protein structures are presented as ribbons.
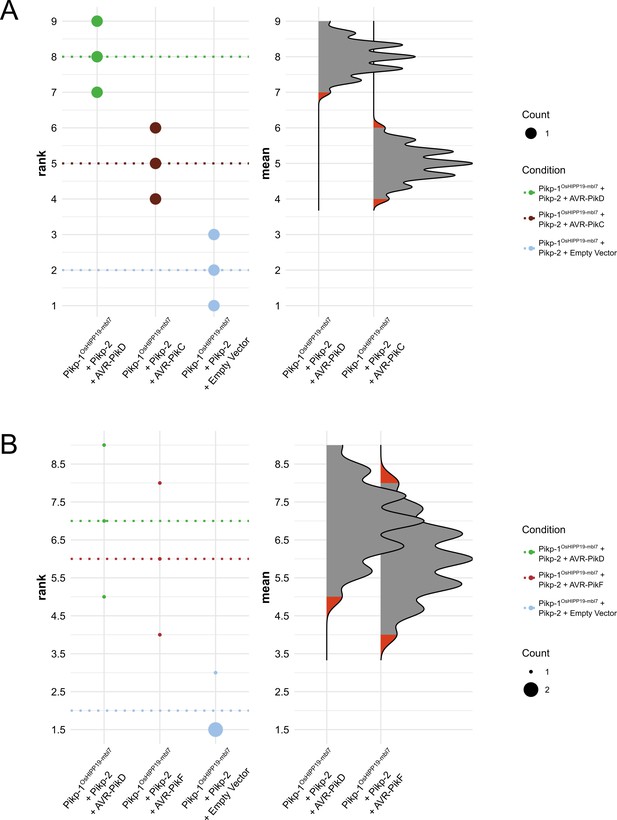
Statistical analysis by estimation methods of the cell death assays presented in Figure 1, for Pikp-1OsHIPP19-mbl7/Pikp-2 with (A) AVR-PikD, AVR-PikC and empty vector, and (B) AVR-PikD, AVR-PikF, and empty vector.
The panel on the left represents the ranked data (dots) for the three replicates of each effector/control, and their corresponding mean (dotted line). The size of the dots is proportional to the number of observations with that specific value. The panel on the right shows the distribution of 1000 bootstrap sample rank means for Pikp-1OsHIPP19-mbl7/Pikp-2/AVR-PikD and Pikp-1OsHIPP19-mbl7/Pikp-2/AVR-PikC (A) or Pikp-1OsHIPP19-mbl7/Pikp-2 (B). The red areas represent the 2.5th and 97.5th percentiles of the distribution. The response of Pikp-1OsHIPP19-mbl7/Pikp-2 to AVR-PikD, AVR-PikC, and AVR-PikF is considered significantly different to the response of Pikp-1OsHIPP19-mbl7/Pikp-2 to the empty vector as the rank means of the latter (dotted line, left panel) falls beyond the red regions of the Pikp-1OsHIPP19-mbl7/Pikp-2/AVR-PikD, Pikp-1OsHIPP19-mbl7/Pikp-2/AVR-PikC, and Pikp-1OsHIPP19-mbl7/Pikp-2/AVR- mean distributions.
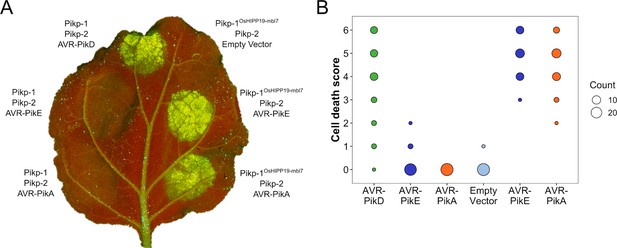
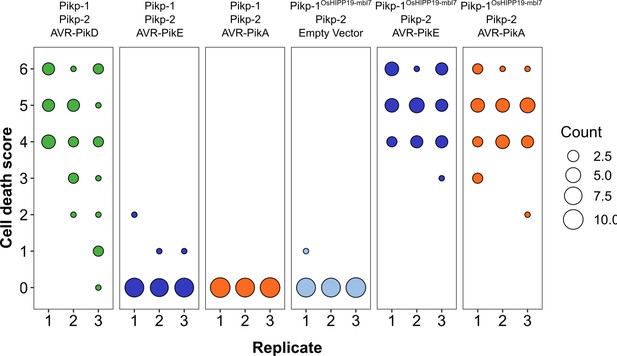
The Pikp-1OsHIPP19-mbl7 chimera responds to AVR-PikE and AVR-PikA.
(A) Representative leaf image showing the Pikp-1OsHIPP19-mbl7 chimera is not autoactive and triggers Pikp-2-dependent cell death in response to AVR-PikE and AVR-PikA. By contrast, Pikp-1 only triggers cell death in response to AVR-PikD, and not AVR-PikE nor AVR-PikA. NLR-mediated responses appear as autofluorescence imaged under UV light. (B) Pikp-mediated response scoring represented as dot plots to summarize 29 repeats of the experiment shown in (A) across three independent experiments (Materials and Methods, Figure 1—figure supplement 11). Fluorescence intensity is scored as stated in Figure 1.
-
Figure 1—figure supplement 10—source data 1
Cell death scores used for dot plots in panel B.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig1-figsupp10-data1-v1.zip
Pikp-mediated response scoring is represented as dot plots, subdivided by replicate, for repeats of the experiment presented in Figure 1—figure supplement 10a.
Replicates 1 and 3 consisted of 10 repeats for each sample, and replicate 2 consisted of 9 repeats for each sample. Fluorescence intensity is scored as described in Figure 1. Scores from the three replicates were combined and represented as the dot plot in Figure 1—figure supplement 10b.
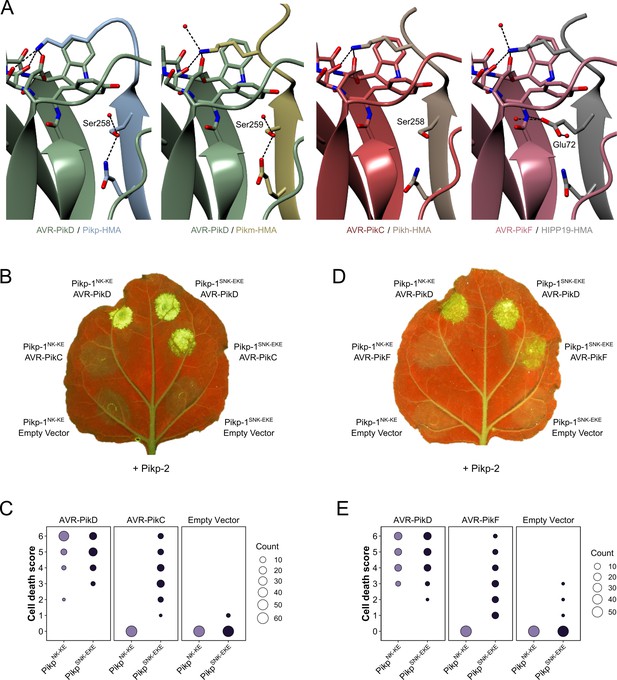
Structure-guided mutagenesis of Pikp-1 expands response to previously unrecognized AVR-Pik effector variants.
(A) Comparison of the crystal structures of AVR-Pik effector variants in complex with Pik-HMA domains (PDB entries 6G10, 6FU9, and 7A8X) and AVR-PikF in complex OsHIPP19 (PDB entry 7B1I) suggests the addition of an S258E mutation to the NK-KE mutations described previously (De la Concepcion et al., 2019) could introduce new contacts across the protein:protein interface. Protein structures are represented as ribbons with relevant side chains displayed as cylinders. Dashed lines indicate hydrogen bonds. Relevant water molecules are represented as red spheres. (B) The PikpSNK-EKE mutant gains response to AVR-PikC (right, middle) where no response is observed for PikpNK-KE (left, middle). Further, the PikpSNK-EKE mutant is not autoactive (right, bottom) and retains response to AVR-PikD (right, top). All infiltration spots contain Pikp-2. (C) Pikp-mediated response scoring represented as dot plots to summarize 60 repeats of the experiment shown in (B) across three independent experiments (Materials and methods, Figure 2—figure supplement 3a). (D) and (E) as described for (B) and (C) but with AVR-PikF and 57 repeats across three independent experiments (Materials and methods, Figure 2—figure supplement 3b).
-
Figure 2—source data 1
Cell death scores used for dot plots in panel C.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-data1-v1.zip
-
Figure 2—source data 2
Cell death scores used for dot plots in panel E.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-data2-v1.zip
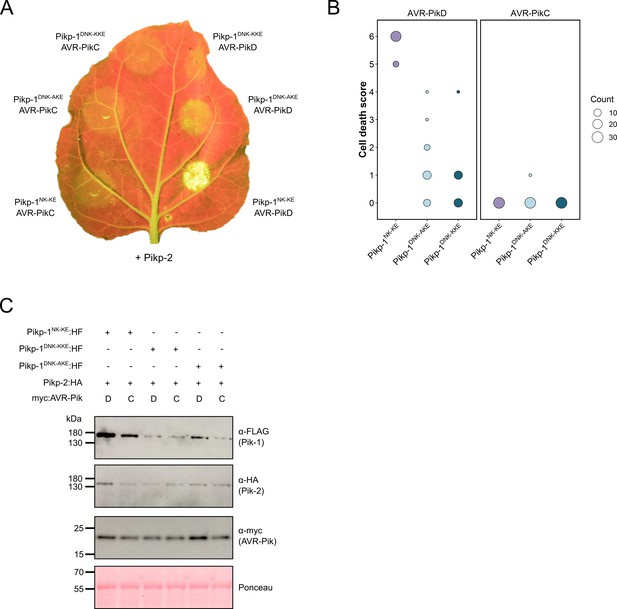
The mutations D224A and D224K mutations in the PikpNK-KE background do not extend response to AVR-PikC.
(A) Neither the Pikp-1DNK-AKE nor the Pikp-1DNK-KKE mutant gains response to AVR-PikC (left, middle and left, top), and response to AVR-PikD is reduced in both mutants (right, middle and right, top). All infiltration spots contain Pikp-2. (B) Pikp-mediated response scoring represented as dot plots to summarize 30 repeats of the experiment shown in (A) across three independent experiments (Materials and methods, Figure 2—figure supplement 2). Fluorescence intensity is scored as stated in Figure 1. (C) Western blots confirming the accumulation of proteins in N. benthamiana. Plant cell lysates were probed for the expression of Pikp-1, Pikp-2, and effector variants, using anti-FLAG, anti-HA, and anti-Myc antiserum, respectively. Total protein extracts were visualized by Ponceau Staining.
-
Figure 2—figure supplement 1—source data 1
Unedited and uncropped blot for panel C, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp1-data1-v1.tiff
-
Figure 2—figure supplement 1—source data 2
Unedited and uncropped blot for panel C, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp1-data2-v1.tiff
-
Figure 2—figure supplement 1—source data 3
Unedited and uncropped blot for panel C, Pik-2 α-HA, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp1-data3-v1.tiff
-
Figure 2—figure supplement 1—source data 4
Unedited and uncropped blot for panel C, Pik-2 α-HA.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp1-data4-v1.tiff
-
Figure 2—figure supplement 1—source data 5
Unedited and uncropped blot for panel C, AVR-Pik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp1-data5-v1.tiff
-
Figure 2—figure supplement 1—source data 6
Unedited and uncropped blot for panel C, AVR-Pik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp1-data6-v1.tiff
-
Figure 2—figure supplement 1—source data 7
Unedited and uncropped blot for panel C, Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp1-data7-v1.tiff
-
Figure 2—figure supplement 1—source data 8
Unedited and uncropped blot for panel C, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp1-data8-v1.tiff
-
Figure 2—figure supplement 1—source data 9
Cell death scores used for dot plots in panel B.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp1-data9-v1.zip
Pikp-mediated response scoring is represented as dot plots, subdivided by replicate, for repeats of the experiment presented in Figure 2—figure supplement 1a.
Each replicate consisted of 10 repeats for each sample. Fluorescence intensity is scored as described in Figure 1. Scores from the three replicates were combined and represented as the dot plot in Figure 2—figure supplement 1b.
Pikp-mediated response scoring represented as dot plots, subdivided by replicate, for repeats of the experiment presented in Figure 2b (A) and 2d (B).
Each replicate consisted of 20 (A) and 19 (B) repeats for each sample. Fluorescence intensity is scored as described in Figure 1. Scores from the three replicates were combined and represented as the dot plots in Figure 2c and e, respectively.
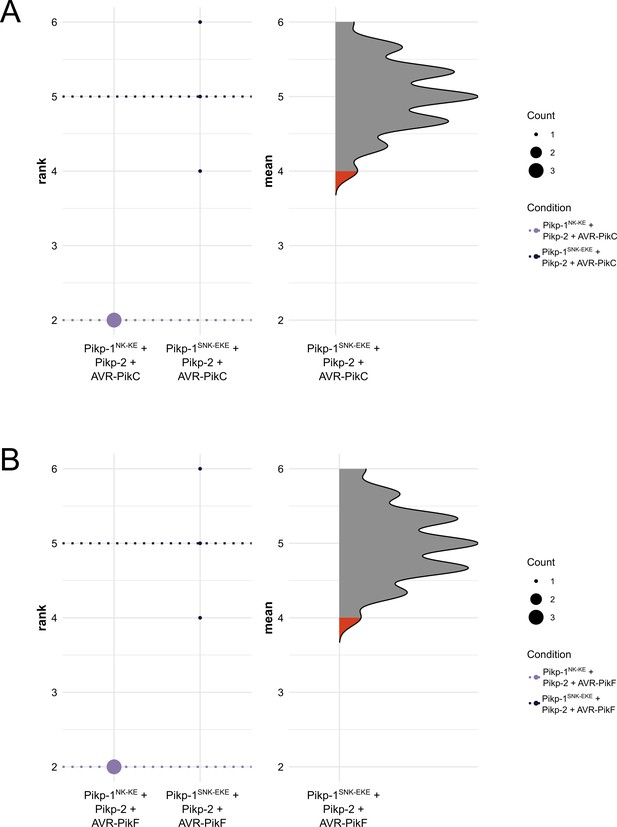
Statistical analysis by estimation methods of the cell death assays presented in Figure 2, for Pikp-1NK-KE/Pikp-2 and Pikp-1SNK-EKE/Pikp-2 with (A) AVR-PikC and (B) AVR-PikF.
The panel on the left represents the ranked data (dots) for the three replicates of each receptor/effector, and their corresponding mean (dotted line). The size of the dots is proportional to the number of observations with that specific value. The panel on the right shows the distribution of 1000 bootstrap sample rank means for Pikp-1SNK-EKE/Pikp-2/AVR-PikC (A) or Pikp-1SNK-EKE/Pikp-2/AVR-PikF (B). The red areas represent the 2.5th and 97.5th percentiles of the distribution. The response of Pikp-1SNK-EKE/Pikp-2 to AVR- AVR-PikC/AVR-PikF is considered significantly different to the response of Pikp-1NK-KE/Pikp-2 to AVR-PikC/AVR-PikF as the rank means of the latter (dotted line, left panel) falls beyond the red regions of the Pikp-1NK-KE/Pikp-2/AVR-PikC and Pikp-1NK-KE/Pikp-2/AVR-PikF mean distributions.
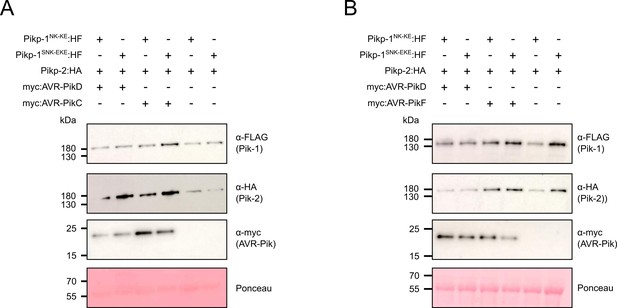
Western blots confirm the accumulation of proteins in N. benthamiana for the cell death assays shown in Figure 2.
(A) Accumulation of proteins for the experiments with AVR-PikC. (B) Accumulation of proteins for the experiments with AVR-PikF. Plant cell lysates were probed for the expression of Pikp-1NK-KE/Pikp-1SNK-EKE, Pikp-2, and AVR-Pik effector variants using anti-FLAG, anti-HA, and anti-Myc antiserum, respectively. Total protein extracts were visualized by Ponceau Staining.
-
Figure 2—figure supplement 5—source data 1
Unedited and uncropped blot for panel A, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data1-v1.tiff
-
Figure 2—figure supplement 5—source data 2
Unedited and uncropped blot for panel A, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data2-v1.tiff
-
Figure 2—figure supplement 5—source data 3
Unedited and uncropped blot for panel A, Pik-2 α-HA, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data3-v1.tiff
-
Figure 2—figure supplement 5—source data 4
Unedited and uncropped blot for panel A, Pik-2 α-HA.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data4-v1.tiff
-
Figure 2—figure supplement 5—source data 5
Unedited and uncropped blot for panel A, AVR-Pik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data5-v1.tiff
-
Figure 2—figure supplement 5—source data 6
Unedited and uncropped blot for panel A, AVR-Pik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data6-v1.tiff
-
Figure 2—figure supplement 5—source data 7
Unedited and uncropped blot for panel A.
Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data7-v1.tiff
-
Figure 2—figure supplement 5—source data 8
Unedited and uncropped blot for panel A, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data8-v1.tiff
-
Figure 2—figure supplement 5—source data 9
Unedited and uncropped blot for panel B, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data9-v1.tiff
-
Figure 2—figure supplement 5—source data 10
Unedited and uncropped blot for panel B, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data10-v1.tiff
-
Figure 2—figure supplement 5—source data 11
Unedited and uncropped blot for panel B, Pik-2 α-HA, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data11-v1.tiff
-
Figure 2—figure supplement 5—source data 12
Unedited and uncropped blot for panel B, Pik-2 α-HA.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data12-v1.tiff
-
Figure 2—figure supplement 5—source data 13
Unedited and uncropped blot for panel B, AVR-Pik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data13-v1.tiff
-
Figure 2—figure supplement 5—source data 14
Unedited and uncropped blot for panel B, AVR-Pik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data14-v1.tiff
-
Figure 2—figure supplement 5—source data 15
Unedited and uncropped blot for panel B, Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data15-v1.tiff
-
Figure 2—figure supplement 5—source data 16
Unedited and uncropped blot for panel B, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp5-data16-v1.tiff
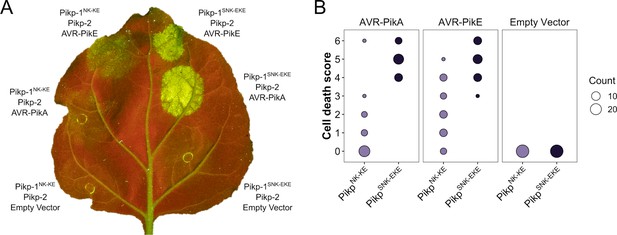
Pikp-1SNK-EKE chimera responds to AVR-PikE and AVR-PikA.
(A) Representative leaf image showing Pikp-1SNK-EKE is not autoactive and triggers Pikp-2-dependent cell death in response to AVR-PikE and AVR-PikA. As previously published (De la Concepcion et al., 2019), Pikp-1NK-KE also triggers cell death in response to AVR-PikE and AVR-PikA in a Pikp-2-dependent manner. Nucleotide-binding and leucine-rich repeat (NLR)-mediated responses appear as autofluorescence imaged under UV light. (B) Pikp-mediated response scoring represented as dot plots to summarize 29 repeats of the experiment shown in (A) across three independent experiments (Materials and methods, Figure 2—figure supplement 7). Fluorescence intensity is scored as stated in Figure 1.
-
Figure 2—figure supplement 6—source data 1
Cell death scores used for dot plots in panel B.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp6-data1-v1.zip
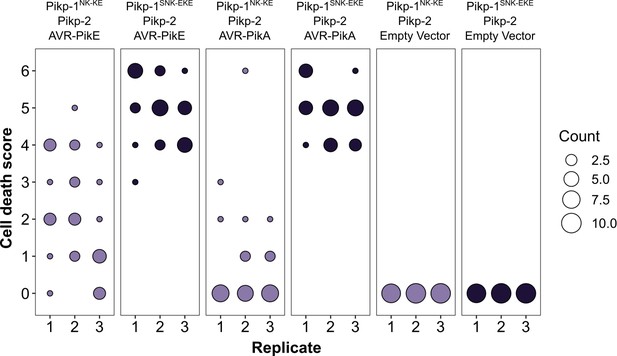
Pikp-mediated response scoring is represented as dot plots, subdivided by replicate, for repeats of the experiment presented in Figure 2—figure supplement 6a.
Replicates 2 and 3 consisted of 10 repeats for each sample, and replicate 1 consisted of 9 repeats for each sample. Fluorescence intensity is scored as described in Figure 1. Scores from the three replicates were combined and represented as the dot plot in Figure 2—figure supplement 6b.
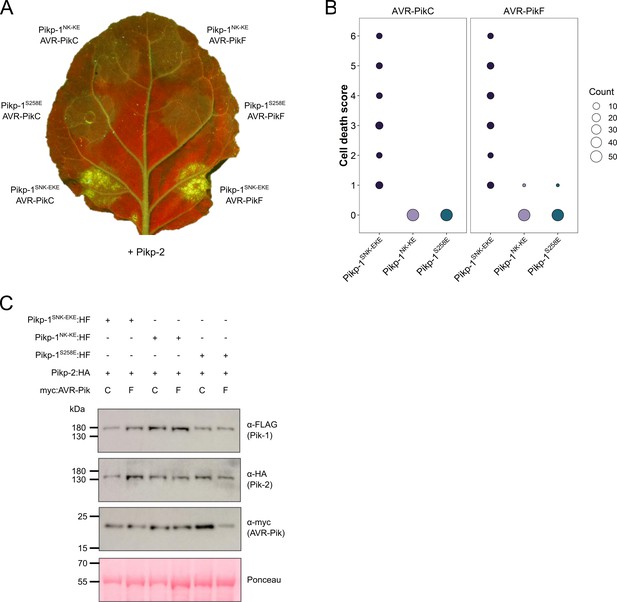
The Pikp S258E mutation alone does not extend response to AVR-PikC or AVR-PikF.
(A) The Pikp-1S258E mutant does not gain response to AVR-PikC (left, middle) or AVR-PikF (right, middle). All infiltration spots contain Pikp-2. (B) Pikp-mediated response scoring represented as dot plots to summarize 54 repeats of the experiment shown in (A) across three independent experiments (Materials and methods, Figure 2—figure supplement 9). Fluorescence intensity is scored as stated in Figure 1. (C) Western blots confirming the accumulation of proteins in N. benthamiana. Plant cell lysates were probed for the expression of Pikp-1, Pikp-2, and effector variants, using anti-FLAG, anti-HA, and anti-Myc antiserum, respectively. Total protein extracts were visualized by Ponceau Staining.
-
Figure 2—figure supplement 8—source data 1
Unedited and uncropped blot for panel C, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp8-data1-v1.zip
-
Figure 2—figure supplement 8—source data 2
Unedited and uncropped blot for panel C, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp8-data2-v1.tiff
-
Figure 2—figure supplement 8—source data 3
Unedited and uncropped blot for panel C, Pik-2 α-HA, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp8-data3-v1.tiff
-
Figure 2—figure supplement 8—source data 4
Unedited and uncropped blot for panel C, Pik-2 α-HA.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp8-data4-v1.tiff
-
Figure 2—figure supplement 8—source data 5
Unedited and uncropped blot for panel C, AVR-Pik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp8-data5-v1.tiff
-
Figure 2—figure supplement 8—source data 6
Unedited and uncropped blot for panel C, AVR-Pik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp8-data6-v1.tiff
-
Figure 2—figure supplement 8—source data 7
Unedited and uncropped blot for panel C, Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp8-data7-v1.tiff
-
Figure 2—figure supplement 8—source data 8
Unedited and uncropped blot for panel C, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp8-data8-v1.tiff
-
Figure 2—figure supplement 8—source data 9
Cell death scores used for dot plots in panel B.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig2-figsupp8-data9-v1.csv
Pikp-mediated response scoring is represented as dot plots, subdivided by replicate, for repeats of the experiment presented in Figure 2—figure supplement 8a.
The three replicates consisted of 18, 20, and 16 repeats for each sample, respectively. Fluorescence intensity is scored as described in Figure 1. Scores from the three replicates were combined and represented as the dot plot in Figure 2—figure supplement 8b, respectively.
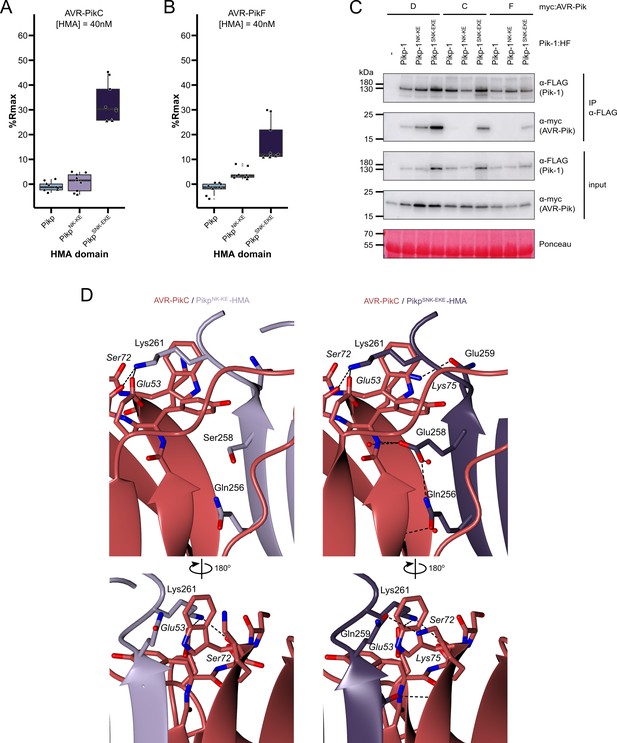
The SNK-EKE triple mutation extends Pikp-1 binding to AVR-PikC and AVR-PikF in vitro and in planta by facilitating new contacts across the protein:protein interface.
Boxplots showing the %Rmax observed for the interactions between AVR-PikC (A) or AVR-PikF (B), both at 40 nM injection concentration, and each of Pikp-HMA, Pikp-HMANK-KE, and Pikp-HMASNK-EKE. %Rmax is the percentage of the theoretical maximum response, assuming a 2:1 binding model (as previously observed for Pikp-HMA proteins). The centre line of the box represents the median and the box limits are the upper and lower quartiles. The whiskers extend to the smallest value within Q1 − 1.5 X the interquartile range (IQR) and the largest value within Q3 +1.5 X IQR. Individual data points are represented as black shapes. The experiment was repeated three times, with each experiment consisting of three technical replicates. Data for 4 nM and 100 nM effector injection concentrations are shown in Figure 3—figure supplement 1. (C) Western blots following co-immunoprecipitation show that the Pikp-1SNK-EKE chimera binds to tested AVR-Pik effector variants in N. benthamiana. Plant cell lysates were probed for the expression of Pikp-1/Pikp-1NK-KE/Pikp-1SNK-EKE and AVR-Pik effector variants using anti-FLAG and anti-Myc antiserum, respectively. Total protein extracts were visualized by Ponceau Staining. (D) The crystal structure of the Pikp-HMASNK-EKE/AVR-PikC complex (PDB entry 7QPX) reveals additional hydrogen bonds at the protein:protein interfaces compared to Pikp-HMANK-KE/AVR-PikC (PDB entry 7A8W). Protein structures are represented as ribbons with relevant side chains displayed as cylinders. Dashed lines indicate hydrogen bonds. Relevant water molecules are represented as red spheres.
-
Figure 3—source data 1
Unedited and uncropped blot for Figure 3C, IP Pik-1 α-FLAG, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data1-v1.tiff
-
Figure 3—source data 2
Unedited and uncropped blot for Figure 3C, IP Pik-1 α-FLAG, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data2-v1.tiff
-
Figure 3—source data 3
Unedited and uncropped blot for Figure 3C, IP Pik-1 α-FLAG, AVRPik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data3-v1.tiff
-
Figure 3—source data 4
Unedited and uncropped blot for Figure 3C, IP Pik-1 α-FLAG, AVRPik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data4-v1.tiff
-
Figure 3—source data 5
Unedited and uncropped blot for Figure 3C, input, Pik-1 α-FLAG, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data5-v1.tiff
-
Figure 3—source data 6
Unedited and uncropped blot for Figure 3C, input, Pik-1 α-FLAG.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data6-v1.tiff
-
Figure 3—source data 7
Unedited and uncropped blot for Figure 3C, input, AVRPik α-myc, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data7-v1.tiff
-
Figure 3—source data 8
Unedited and uncropped blot for Figure 3C, input, AVRPik α-myc.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data8-v1.tiff
-
Figure 3—source data 9
Unedited and uncropped blot for Figure 3C, Ponceau stain, with relevant bands labeled.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data9-v1.tiff
-
Figure 3—source data 10
Unedited and uncropped blot for Figure 3C, Ponceau stain.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data10-v1.tiff
-
Figure 3—source data 11
Surface plasmon resonance (SPR) data used for box plots in panel A.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data11-v1.zip
-
Figure 3—source data 12
Surface plasmon resonance (SPR) data used for box plots in panel B.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig3-data12-v1.zip
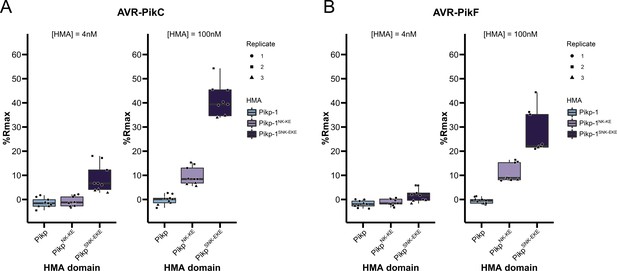
Boxplots showing the %Rmax observed for the interactions between AVR-PikC (A) or AVR-PikF (B), both at 4 nM and 100 nM injection concentrations, and each of Pikp-HMA, Pikp-HMANK-KE, and Pikp-HMASNK-EKE.
%Rmax is the percentage of the theoretical maximum response, assuming a 2:1 binding model (as previously observed for Pikp-HMA proteins, see Materials and methods).The center line of the box represents the median and the box limits are the upper and lower quartiles. The whiskers extend to the smallest value within Q1 − 1.5 Å ~the interquartile range (IQR) and the largest value within Q3 + 1.5 Å ~IQR. Individual data points are represented as black shapes. The experiment was repeated three times, with each experiment consisting of three technical replicates.
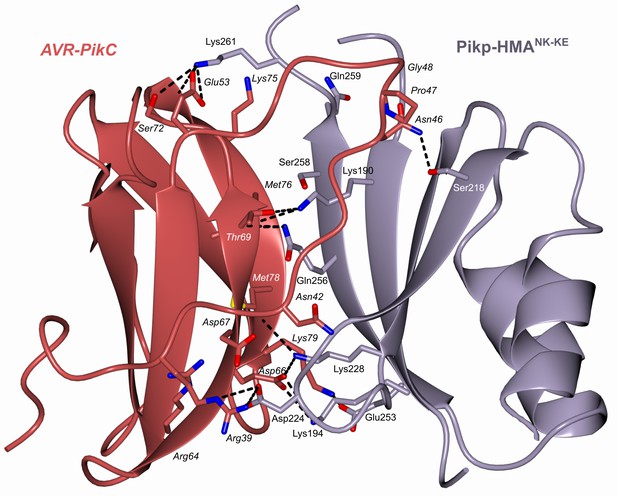
Schematic representation of the crystal structure of the complex formed between Pikp-HMANK-KE and AVR-PikC (PDB entry 7A8W).
The overall structure is similar to other Pik-HMA/AVR-Pik complexes. Amino acid residues forming key contacts at the interface are labeled, including Asp67 which distinguishes AVR-PikC from AVR-PikE.
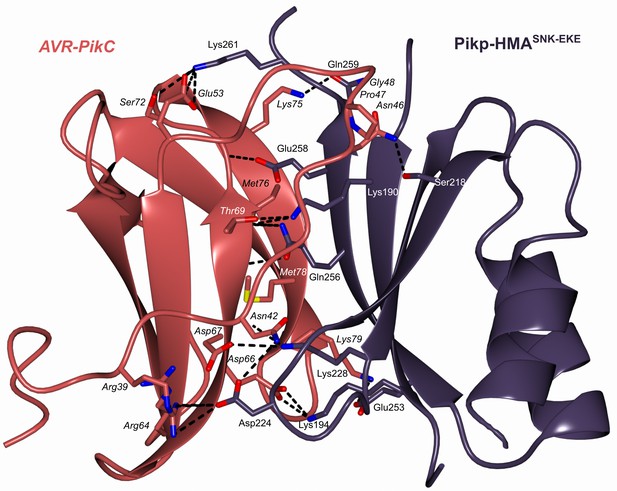
Schematic representation of the crystal structure of the complex formed between Pikp-HMASNK-EKE and AVR-PikC (PDB entry 7QPX).
The overall architecture of the complexes is similar to other Pik-HMA/AVR-Pik structures. Amino acid residues forming key contacts at the interface are labeled, including Asp67 which distinguishes AVR-PikC from AVR-PikE.
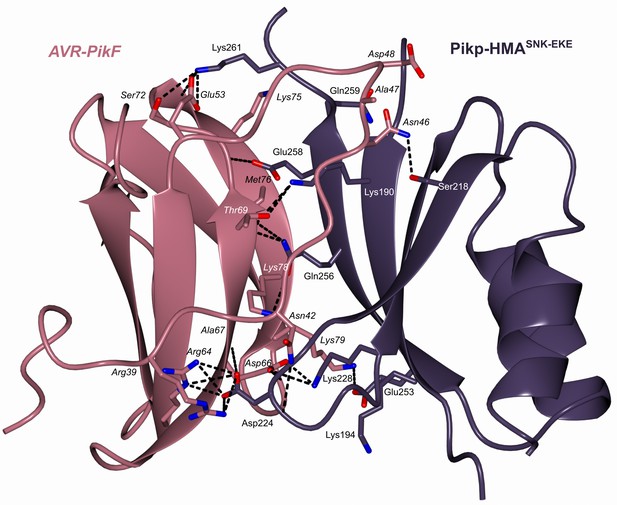
Schematic representation of the crystal structure of the complex formed between Pikp-HMASNK-EKE and AVR-PikF (PDB entry 7QZD).
The overall architecture of the complexes is similar to other Pik-HMA/AVR-Pik structures. Amino acid residues forming key contacts at the interface are labeled, including Lys78 which distinguishes AVR-PikF from AVR-PikA.
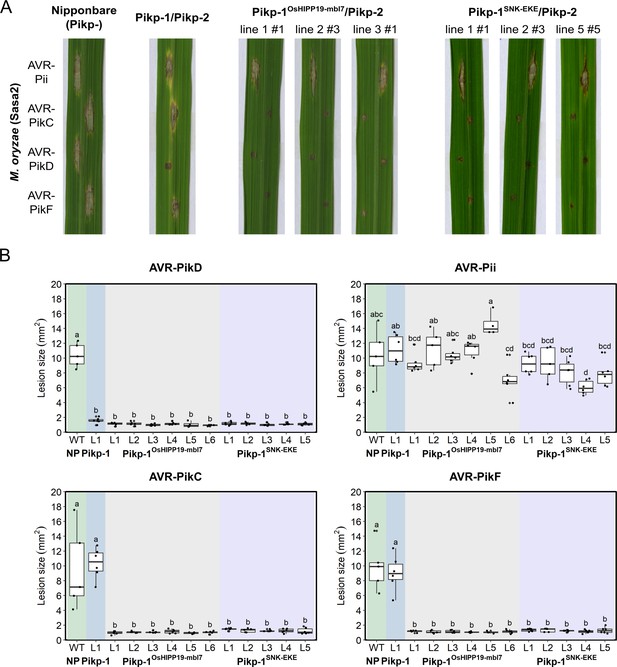
Transgenic rice plants carrying the Pikp-1OsHIPP19mbl7 chimera or the Pikp-1SNK-EKE mutation show extended resistance to Magnaporthe oryzae carrying AVR-PikC or AVR-PikF compared to Pikp-1 wild-type.
(A) Example leaves from pathogenicity assays of wild-type O. sativa cv. Nipponbare and three transgenic lines of O. sativa cv. Nipponbare expressing Pikp-1/Pikp-2, Pikp-1OsHIPP19mbl7/Pikp-2, or Pikp-1SNK-EKE/Pikp-2 challenged with M. oryzae Sasa2 transformed with AVR-Pii, AVR-PikC, AVR-PikD, or AVR-PikF. The T1 generation seedlings were used for the inoculation test. Wild-type O. sativa cv. Nipponbare (recipient) is susceptible to all M. oryzae Sasa2 transformants (left), while the Pikp-1/Pikp-2 transformant is only resistance to M. oryzae Sasa2 transformed with AVR-PikD (no development of disease lesions). The Pikp-1OsHIPP19mbl7/Pikp-2 or Pikp-1SNK-EKE/Pikp-2 plants show resistance to M. oryzae Sasa2 transformed with AVR-PikC, AVR-PikD or AVR-PikF but not AVR-Pii. (B) Disease lesion sizes (determined using ImageJ) are represented as box plots. L1-L6 and L1-L5 represent data from rice lines as shown in Figure 4—figure supplements 2–4. The centre line of the box represents the median and the box limits are the upper and lower quartiles. The whiskers extend to the smallest value within Q1 − 1.5 X the interquartile range (IQR) and the largest value within Q3 + 1.5 X IQR. Individual measurements are represented as black dots. Leaf images corresponding to each of the data points presented in the box plots are shown in Figure 4—figure supplements 2–4. Lowercase letters represent statistically significant differences (p<0.05, ANOVA with post-hoc Tukey HSD test). RT-PCR confirmed expression of transgenes in (A) is shown in Figure 4—figure supplement 1.
-
Figure 4—source data 1
Lesion size data for pathogen growth on rice plants.
- https://cdn.elifesciences.org/articles/81123/elife-81123-fig4-data1-v1.zip
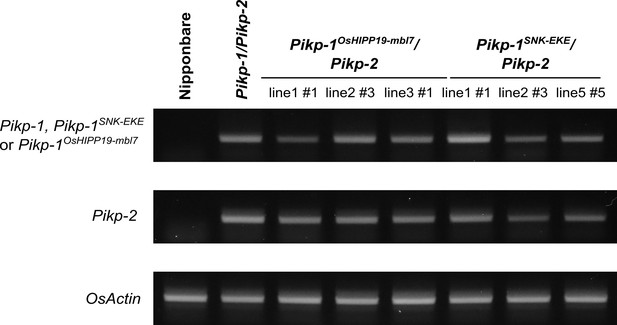
RT-PCR to confirm transgene expression of Pikp-1 (or engineered Pikp-1 variants) and Pikp-2 for the individuals shown in Figure 4.
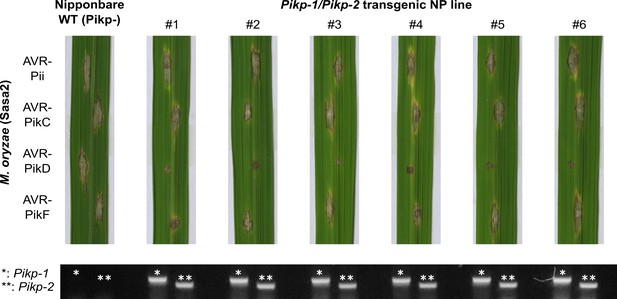
Pathogenicity assays in the T1 progenies derived from a Pikp-1/Pikp-2 T0 transgenic line of O. sativa cv. Nipponbare against M. oryzae Sasa2 transformed with AVR-Pii, AVR-PikC, AVR-PikD, or AVR-PikF.
Images of leaves from different T1 lines were taken 7 days after inoculation. Gel images show PCR confirmation of transgenes. The plants show susceptibility to all M. oryzae strains in the recipient line (WT) and resistance only to AVR-PikD in the transgenics.
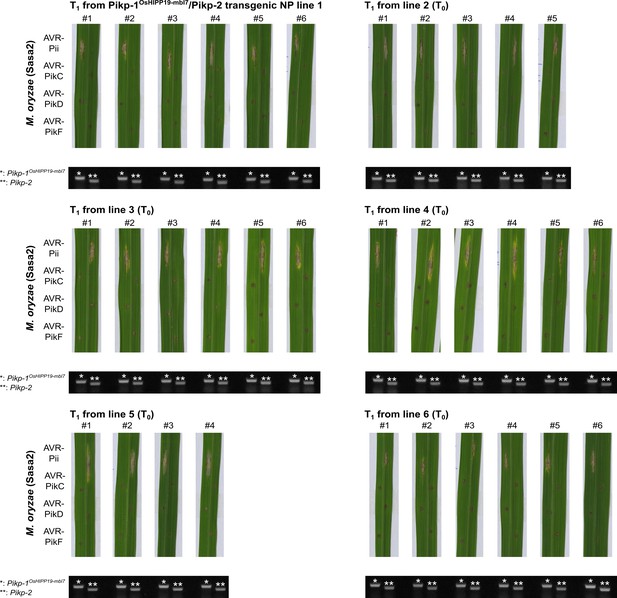
Pathogenicity assays in the T1 progenies derived from six independent Pikp-1OsHIPP19-mbl7/Pikp-2 T0 transgenic lines of O. sativa cv. Nipponbare against M. oryzae Sasa2 transformed with AVR-Pii, AVR-PikC, AVR-PikD, or AVR-PikF.
Images of leaves from different T1 lines were taken 7 days after inoculation. Gel images show PCR confirmation of transgenes. The plants show resistance to all M. oryzae strains carrying AVR-Pik effectors, but are susceptible to M. oryzae Sasa2 carrying AVR-Pii.
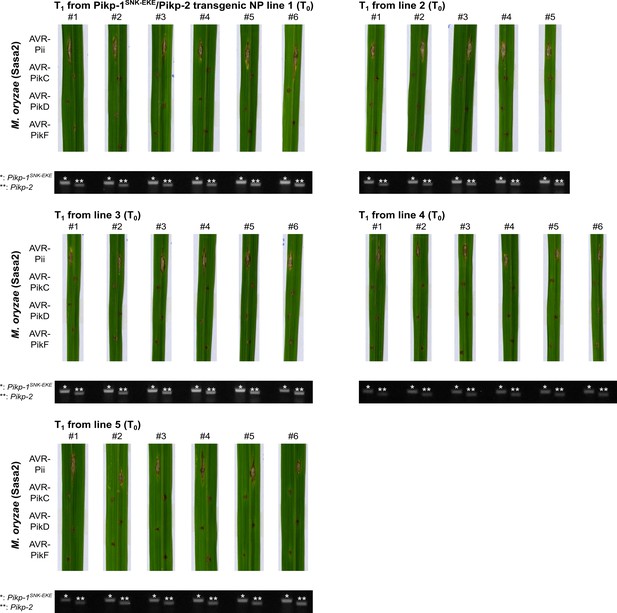
Pathogenicity assays in the T1 progenies derived from five independent Pikp-1SNK-EKE/Pikp-2 T0 transgenic lines of O. sativa cv. Nipponbare against M. oryzae Sasa2 transformed with AVR-Pii, AVR-PikC, AVR-PikD, or AVR-PikF.
Images of leaves from different T1 lines were taken 7 days after inoculation. Gel images show PCR confirmation of transgenes. The plants show resistance to all M. oryzae strains carrying AVR-Pik effectors, but are susceptible to M. oryzae Sasa2 carrying AVR-Pii.
Additional files
-
Supplementary file 1
Table S1.
X-ray data collection and refinement statistics for Pikp-HMANK-KE/AVR-PikC (PDB entry 7A8W), Pikp-HMASNK-EKE/AVR-PikC (PDB entry 7QPX), and Pikp-HMASNK-EKE/AVR-PikF (PDB entry 7QZD).
- https://cdn.elifesciences.org/articles/81123/elife-81123-supp1-v1.docx
-
Supplementary file 2
Table S2.
Summary of interface analysis by QtPISA for Pikp-HMANK-KE/AVR-PikC (PDB entry 7A8W), Pikp-HMASNK-EKE/AVR-PikC (PDB entry 7QPX), and Pikp-HMASNK-EKE/AVR-PikF (PDB entry 7QZD). Protein chains used for the analysis in each complex (as defined in the PDB entries) are: PikpNK-KE:AVR-PikC (E and F); PikpSNK-EKE:AVR-PikC (E and F); PikpSNK-EKE:AVR-PikF (F and G).
- https://cdn.elifesciences.org/articles/81123/elife-81123-supp2-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/81123/elife-81123-mdarchecklist1-v1.pdf





