Embryo-derive TNF promotes decidualization via fibroblast activation
Abstract
Decidualization is a process in which endometrial stromal fibroblasts differentiate into specialized secretory decidual cells and essential for the successful establishment of pregnancy. The underlying mechanism during decidualization still remains poorly defined. Because decidualization and fibroblast activation share similar characteristics, this study was to examine whether fibroblast activation is involved in decidualization. In our study, fibroblast activation-related markers are obviously detected in pregnant decidua and under in vitro decidualization. ACTIVIN A secreted under fibroblast activation promotes in vitro decidualization. We showed that arachidonic acid released from uterine luminal epithelium can induce fibroblast activation and decidualization through PGI2 and its nuclear receptor PPARδ. Based on the significant difference of fibroblast activation-related markers between pregnant and pseudopregnant mice, we found that embryo-derived TNF promotes CPLA2α phosphorylation and arachidonic acid release from luminal epithelium. Fibroblast activation is also detected under human in vitro decidualization. Similar arachidonic acid-PGI2-PPARδ-ACTIVIN A pathway is conserved in human endometrium. Collectively, our data indicate that embryo-derived TNF promotes CPLA2α phosphorylation and arachidonic acid release from luminal epithelium to induce fibroblast activation and decidualization.
Editor's evaluation
The authors provide novel evidence for a connection between fibroblast activation and eutherian stromal decidualization. This important work substantially advances our understanding of decidua biology and its contribution to pregnancy. The authors are using solid evidence to support the findings.
https://doi.org/10.7554/eLife.82970.sa0Introduction
Endometrial stromal fibroblasts undergo decidualization to form specialized secretory decidual cells. Decidualization is characterized by significant proliferation, differentiation, and epithelial transition of endometrial stromal cells (Gellersen and Brosens, 2014; Tan et al., 2004). In mice, decidualization occurs only after the onset of implantation. However, decidualization can also be induced by artificial stimuli (McConaha et al., 2011). In contrast to mice, human decidualization does not require embryo implantation and is driven by the rise of postovulatory progesterone and local cyclic adenosine monophosphate (cAMP) from endometrial stromal cells (Gellersen and Brosens, 2014). Although decidualization is essential to the successful establishment of pregnancy, the underlying mechanism during decidualization is still poorly defined. Accumulating evidence indicates that decidualization and fibroblast activation share similar characteristics. During primate decidualization,αSMA increases at the initial stage and decreases at the final differentiation stage (Kim et al., 1999). αSMA is also strongly expressed in rat decidual cells (Venuto et al., 2008). αSMA expression in interstitial fibroblasts during pregnancy correlates with the onset of the decidual process (Fazleabas et al., 1999). Meanwhile, αSMA is strongly expressed in myofibroblast and often recognized as the marker of fibroblast activation (Fujigaki et al., 2005; Angelini et al., 2020). ATP and uric acid belong to damage-associated molecular patterns (DAMPs), which are released following tissue injury or cell death (Chen and Nuñez, 2010). We recently demonstrated that either ATP or uric acid can induce mouse decidualization (Gu et al., 2020; Zhu et al., 2021). It has been shown that ATP or uric acid can also stimulate fibroblast activation (Dolmatova et al., 2012; Bao et al., 2018).
Fibroblasts are the most numerous cells in connective tissue. Fibroblast activation refers to the process in which dormant quiescent fibroblasts in tissues are stimulated to form functionally active fibroblasts and perform different functions. Fibroblasts synthesize the extracellular matrix of connective tissue and play a key role in maintaining the structural integrity of most tissues (Yoshida, 2020; Tracy et al., 2016; Enzerink and Vaheri, 2011). In healthy and intact tissues, fibroblasts remain a dormant and non-proliferating state. Upon stimulation, dormant fibroblasts acquire contractile properties by inducing the formation of stress fibers, resulting in the formation of myofibroblasts (Pakshir et al., 2020; Tomasek et al., 2002). The myofibroblast is a specialized fibroblast expressing α-smooth muscle actin (α-SMA; Angelini et al., 2020). α-SMA is also a marker of cancer-associated fibroblasts (CAFs; Nurmik et al., 2020). In tumor tissues, activated stromal fibroblasts are called as CAFs and show similar characteristics with myofibroblasts (Shimura, 2021). Under the stimulation of cytokines and growth factors, fibroblasts will transform into myofibroblasts at the initial stage and further differentiate to into functionally fibroblasts. Activated fibroblasts are characterized by expressing α-SMA, FAP, Vimentin, Desmin, S100A4(also called FSP1), Tenascin C (TNC), periostin, and SPARC (Kuzet and Gaggioli, 2016).
In adult tissues, the human endometrium undergoes cyclical shedding and bleeding, scar-free repair and regeneration in subsequent cycles (Martin, 2007; Salamonsen et al., 2021). In the uterus, the mesenchyme accounts for about 30–35% of the main uterine cell types. Six cell types have been identified in human endometrium, including stromal fibroblasts, endothelial cells, macrophages, uNK, lymphocytes, epithelial cells, and smooth muscle cells (Wang et al., 2020). Based on a recent single-cell transcriptomic analysis of human endometrium, stromal cells were the most abundant cell type in the endometrium (Lv et al., 2022b). As mouse blastocyst begins to adhere to the luminal epithelium on day 4 of pregnancy, the activated embryo secretes a series of factors to remodel the stationary fibroblasts through epithelial cells (Li et al., 2015). Whether fibroblast activation is involved in embryo implantation and decidualization is still unknown.
Arachidonic acid (AA) is the biosynthetic precursor of prostaglandins in the cell membrane. CPLA2α is encoded by the Pla2g4a gene and a major provider of AA. Pla2g4a knockout mice show uneven embryo distribution and reduced litter size, suggesting that maternal uterine CPLA2α is critical for successful embryo implantation (Song et al., 2002). Cyclooxygenase COX is the rate-limiting enzyme for prostaglandin synthesis. COX-1 knockout mice are fertile, with only birth defects (Lim and Dey, 1997). COX-2 knockout on C57BL/6 J/129 background mice results in impaired implantation, and decidualization, indicating the important role of COX-2 during embryo implantation and decidualization (Lim and Dey, 1997). Prostaglandin I2 (PGI2) and prostaglandin E2 (PGE2) are the two most abundant prostaglandins at the embryo implantation site (Lim et al., 1999). PGI2 derived from COX-2 and prostacyclin synthase (PGIS) can regulate embryo implantation through peroxisome proliferators-activated receptor δ (PPARδ; Lim et al., 1999). Both embryonic implantation and decidualization are abnormal in PPARδ-/- mice (Wang et al., 2007). Although prostaglandins are required for embryo implantation and decidualization, whether prostaglandins are involved in fibroblast activation during early pregnancy remains unclear.
This study was to analyze whether fibroblast activation is involved in mouse and human decidualization, and why fibrosis does not occur during normal early pregnancy. In this study, the multiple markers of fibroblast activation were detected during mouse in vivo and in vitro decidualization. Embryo-derived TNF was able to induce the phosphorylation of CPLA2α in luminal epithelium, which liberated AA into uterine stroma to promote fibroblast activation through COX2-PGI2-PPARδ pathway for inducing decidualization. The underlying mechanism of fibroblast activation was also conserved during human in vitro decidualization. This study offers novel insights into the function of fibroblasts during embryo implantation and decidualization.
Results
Fibroblast activation is present during mouse early pregnancy
S100A4, αSMA, TNC, and SPARC are well-recognized markers of fibroblast activation (Kuzet and Gaggioli, 2016; Prime et al., 2017). Immunofluorescence and immunohistochemistry showed that TNC, SPARC, and S100A4 were mainly localized in the primary decidual region after embryo implantation. SPARC immunofluorescence was strongly observed in subluminal stromal cells from D4.5 to D5.5. However, TNC immunostaining was strongly detected in subluminal stromal cells on D4, D4.5, and D5, but disappeared on D5.5. S100A4 immunostaining wasn’t detected in mouse uterus on D4 and D4.5 of pregnancy, but detected in subluminal stromal cells at implantation sites on D5 and D5.5 (Figure 1A). Under in vitro decidualization, the protein levels of αSMA, TNC and SPARC were significantly increased by 4.41, 3.25, and 2.69-folds compared with the control group, respectively (Figure 1B). These results suggested that fibroblast activation should be present under in vivo and in vitro decidualization.
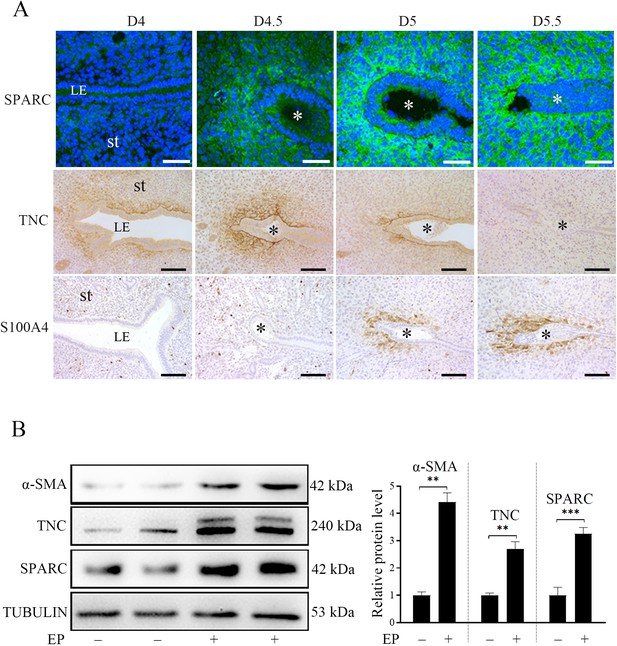
The protein localization and levels of markers of fibroblast activation in mouse uteri during early pregnancy.
(A) Immunofluorescence of SPARC and immunohistochemistry of TNC and S100A4 in mouse uteri on D4 (n=5), D4.5 (n=5), D5 (n=5), and D5.5 (n=5) of pregnancy. LE, luminal epithelium; St, stroma; * Embryo. Scale bar, 50 μm. (B) Western blot analysis of α-SMA, SPARC, TNC protein level under in vitro decidualization (EP) for 24 hr. *, p<0.05; **, p<0.01; ***, p<0.001.
-
Figure 1—source data 1
Raw data of all western blots from Figure 1.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig1-data1-v2.zip
-
Figure 1—source data 2
Complete and uncropped membranes of all western blots from Figure 1.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig1-data2-v2.zip
Fibroblast activation promotes decidualization by secreting ACTIVIN A
BMP2 and WNT4 are essential to mouse decidualization (Lee et al., 2007; Li et al., 2007). E2F8 and CYCLIN D3 are markers of polyploidy during mouse decidualization (Qi et al., 2015). It has been shown that activated fibroblasts can secrete S100A4, SPARC, TNC, and ACTIVIN A (Kuzet and Gaggioli, 2016; Truffi et al., 2020). In order to examine whether fibroblast activation is involved in mouse decidualization, mouse stromal cells were treated with S100A4, SPARC, TNC, and ACTIVIN A to induce in vitro decidualization, respectively. TNC treatment upregulated the protein levels of WNT4 and CYCLIN D3, but had no obvious effects on BMP2, E2F8, and Prl8a2 (Figure 2A and B). S100A4 treatment upregulated BMP2 and WNT4 protein levels by 1.76- and 1.42-folds, respectively, but had no obvious effects on E2F8, CYCLIN D3, and Prl8a2 (Figure 2C and D). SPARC treatment on mouse stromal cells had no significant effect on decidualization markers (Figure 2E and F). Because ACTIVIN A is secreted under fibroblast activation (Loomans and Andl, 2014), we examined ACTIVIN A protein levels in the uteri on D4 and D4.5 of pregnancy and pseudopregnant (PD4, PD4.5) mice, respectively. The level of ACTIVIN A protein on D4 was 1.75-folds that of PD4 mice, and the level of ACTIVIN A protein on D4.5 was 3.77-folds that of PD4.5 mice. The protein level of ACTIVIN A on D4.5 was 1.88-folds higher than that on D4 (Figure 2G), indicating that the secretion of ACTIVIN A increased after embryo implantation. After mouse stromal cells were treated with ACTIVIN A, all the protein levels of BMP2, WNT4, E2F8, CYCLIN D3, and Prl8a2 were obviously up-regulated 1.35, 2.36, 3.11, 1.90, and 6.29 folds, respectively (Figure 2H, I). Under in vitro decidualization, ACTIVIN A treatment also increased all the protein levels of BMP2, WNT4, E2F8, and CYCLIN D3 by 1.52, 1.43, 3.89, and 1.36 folds, respectively (Figure 2J). These results suggest a positive correlation between ACTIVIN A and decidualization.
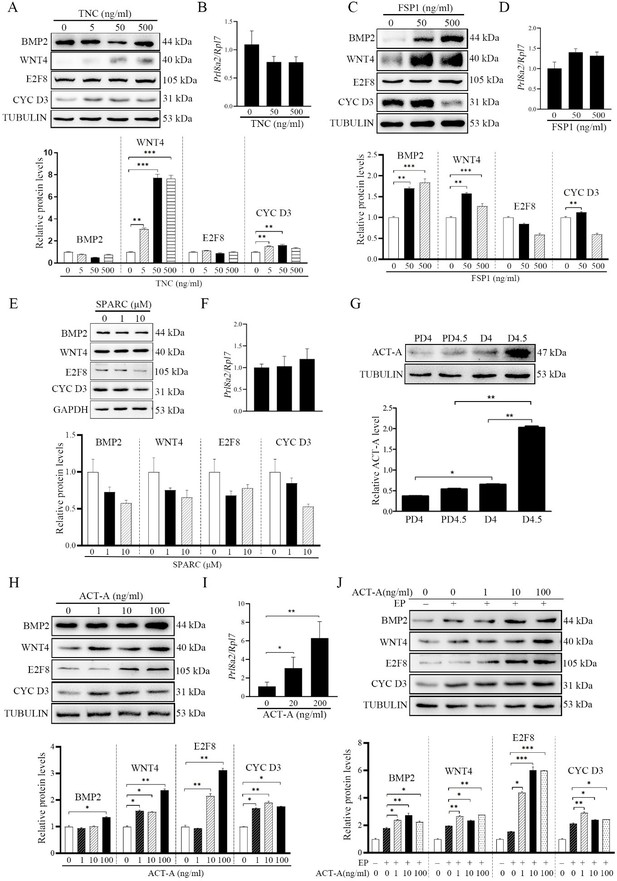
Fibroblast activation promotes decidualization by secreting ACTIVIN A.
(A) Western blot analysis on the effects of TNC on decidualization markers (BMP2, WNT4, E2F8 and CYCLIN D3) after stromal cells were treatment with TNC for 72 hr. (B) QPCR analysis of Prl8a2 mRNA level after mouse stromal cells were treated with TNC for 72 hr. (C) Western blot analysisof the effects of S100A4 on decidualization markers after stromal cells were treated with S100A4 for 72 hr. (D) QPCR analysis of Prl8a2 mRNA level after mouse stromal cells were treated with S100A4 for 72 hr. (E) Western blot analysis on the effects after stromal cells were treated with SPARC for 72 hr. (F) QPCR analysis of Prl8a2 mRNA level after mouse stromal cells were treated SPARC for 72 hr. (G) Western blot analysis on ACTIVIN A protein levels in mouse uteri on D4, D4.5, PD4, and PD4.5, respectively. (H) Western blot analysis on the effects of ACTIVIN A on decidualization markers after stromal cells were treated with ACTIVIN A for 72 hr. (I) QPCR analysis of Prl8a2 mRNA level after mouse stromal cells were treated with ACTIVIN A for 72 hr. (J) Western blot analysis on the effects of ACTIVIN A on decidualization markers after stromal cells were treated with ACTIVIN A for 48 hr under in vitro decidualization. EP, 17β-estradiol+progesterone. All data were is presented as means ± SD. *, p<0.05; **, p<0.01; ***, p<0.001. CYC D3: CYCLIN D3; ACT-A: ACTIVIN A.
-
Figure 2—source data 1
Raw data of all western blots from Figure 2.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig2-data1-v2.zip
-
Figure 2—source data 2
Complete and uncropped membranes of all western blots from Figure 2.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig2-data2-v2.zip
PGI2 promotes fibroblast activation and decidualization through PPARδ pathway
Although we just showed the presence of fibroblast activation during decidualization, what initiates fibroblast activation is still unknown. A previous study indicated that PGE2 and PGI2 are the most abundant prostaglandins at implantation sites in mouse uterus (Lim et al., 1999). Both PGE2 and PGI2 are essential to mouse decidualization (Lim et al., 1999; Holmes and Gordashko, 1980). When mouse stromal cells were treated with PGE2 in the absence or presence of progesterone (P4), PGE2 had no obvious effects on the protein levels of αSMA, this was despite significant increases in TNC and SPARC (Figure 3A). However, ILOPROST, a PGI2 analog (Huang et al., 2002), up-regulated the protein levels of TNC, αSMA and SPARC (2.65, 1.80, and 1.53 folds). PGI2 membrane receptor (IP) (Namba et al., 1994) was down-regulated by ILOPROST treatment, but peroxisome proliferator-activated receptor δ (PPARδ), the PGI2 nuclear receptor, was significantly increased by ILOPROST (2.54 folds) (Figure 3B). PPARδ agonist GW501516 was also able to upregulate the protein levels of TNC, αSMA and SPARC by 7.77, 2.81, and 2.03 folds, respectively (Figure 3C). Further analysis showed that treatment of stromal cells with 0.1 μM of IP agonist SELEXIPAG (Asaki et al., 2015) increased a-SMA and SPARC levels by 1.39 and 2.26 folds, respectively, but had no obvious effect on TNC level. When stromal cells were treated with 1 or 10 μM of SELEXIPAG, the protein levels of a-SMA and SPARC decreased (Figure 3D). After stromal cells were treated with either ILOPROST or GW501516, decidualization markers (BMP2, WNT4, E2F8, and CYCLIN D3) were also significantly up-regulated by 1.96, 1.81, 1.33, and 1.87, or 1.73, 5.34, 3.73, and 1.91 folds, respectively (Figure 3E and G). The protein levels of WNT4 and CYCLIN D3 were decreased by 10 μM ILOPROST (Figure 3G). The mRNA levels of Prl8a2 were also increased by either ILOPROST or GW501516 (Figure 3F and H). Compared with control, the protein level of ACTIVIN A was obviously upregulated by 0.1 μM ILOPROST (1.48 folds) (Figure 3I). These results showed that PGI2 initiated fibroblast activation and promoted decidualization through the PPARδ pathway.
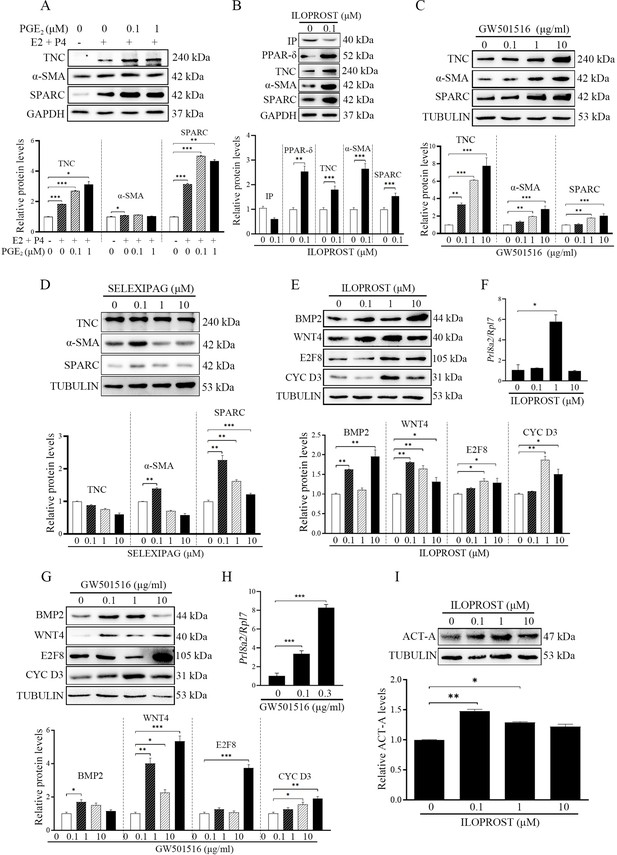
Western blot analysis on effects of prostaglandins on fibroblast activation and decidualization in mouse stromal cells.
(A) The effects of PGE2 on markers of fibroblast activation. (B) The effects of ILOPROST, PGI2 analog, on markers of fibroblast activation after stromal cells was treated with ILOPROST for 12 hr. (C) The effects of GW501516, PPARδ agonist, on markers of fibroblast activation. (D) The effects of SELEXIPA, IP analog, on markers of fibroblast activation. (E) The effects of ILOPROST on decidualization markers. (F) QPCR analysis of Prl8a2 mRNA level after mouse stromal cells were treated with ILOPROST for 48 hr. (G) The effects of GW501516 on decidualization markers. (H) QPCR analysis of Prl8a2 mRNA level after mouse stromal cells were treated with GW501516 for 48 h. (I) The effects of ILOPROST on ACTIVIN A protein levels after stromal cells were treated with ILOPROST for 24 hr. CYC D3: CYCLIN D3; ACT-A: ACTIVIN A. *, p<0.05; **, p<0.01; ***, p<0.001.
-
Figure 3—source data 1
Raw data of all western blots from Figure 3.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig3-data1-v2.zip
-
Figure 3—source data 2
Complete and uncropped membranes of all western blots from Figure 3.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig3-data2-v2.zip
Arachidonic acid induces the fibroblasts activation and promotes decidualization through stimulating Activin A secretion
AA is the precursor of prostaglandin biosynthesis and can be liberated from membrane lipids through the phosphorylation of CPLA2α (Simmons et al., 2004). CPLA2α is significantly expressed in the luminal epithelium in mouse uterus during peri-implantation period and essential for mouse embryo implantation (Song et al., 2002), suggesting that AA should be released from luminal epithelium. After stromal cells were treated with AA, the markers of fibroblast activation (TNC, αSMA, and SPARC) were significantly up-regulated (1.63, 16.00. and 31.20 folds) (Figure 4A). Treatment with AA also upregulated the protein levels of decidualization markers (BMP2 by 1.25-folds, WNT4 by 1.27-folds, E2F8 by 15.78-folds and CYCLIN D3 by 6.31-folds) (Figure 4B) and the mRNA level of Prl8a2 (another marker for mouse decidualization) (Figure 4D). Under in vitro decidualization, AA treatment also significantly increased the decidualization markers (BMP2 by 3.54-folds, WNT4 by 1.81-folds, E2F8 by 3.09-folds and CYCLIN D3 by 2.68-folds; Figure 4C). These results indicated that AA might promote decidualization through fibroblast activation. Additionally, treatment with AA distinctly stimulated the protein levels of COX-2, PGIS, and PPARδ (10.94, 1.99, and 53.03 folds), but had no obvious effects on PGES and IP (Figure 4E). The stimulation of AA on PGIS and PPARδ was abrogated by COX-2 inhibitor NS398 (Figure 4F). The induction of AA on Prl8a2 mRNA levels was also suppressed by either COX-2 inhibitor NS398 or PPARδ antagonist GSK0660 (Figure 4H and I). AA also significantly induced the protein level of ACTIVIN A and the mRNA level of Inhba (encoded for ACTIVIN A; Figure 4G and J). Furthermore, the induction of AA on Prl8a2 expression was abrogated by either ACTIVIN A inhibitor (SB431542) or Inhba siRNA (Figure 4K to M). These results suggested that AA induced fibroblast activation and promoted decidualization through ACTIVIN A.
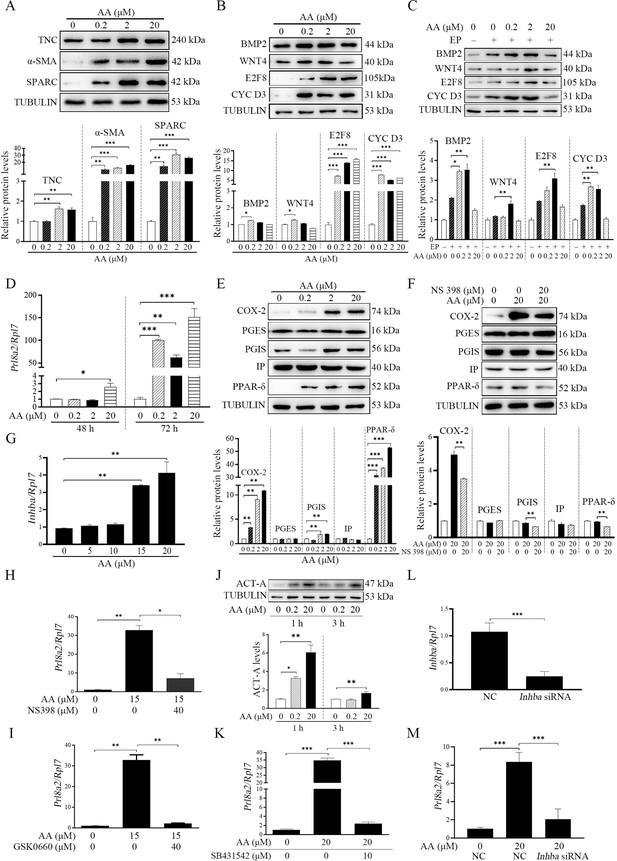
Effects of AA on fibroblast activation and decidualization through PGI-PPARδ-ACTIVIN A pathway.
(A) Western blot analysis on effects of AA on markers of fibroblast activation after stromal cells were treated with AA for 6 hr. (B) Western blot analysis on effects of AA on decidualization markers after stromal cells were treated with AA for 48 hr. (C) Western blot analysis on effects of AA on decidualization markers after stromal cells were treated with AA for 48 hr under in vitro decidualization. EP, 17β-estradiol+progesterone. (D) QPCR analysis of Prl8a2 mRNA level after stromal cells were treated with AA for 72 hr. (E) Western blot analysis on effects of AA on COX2, PGES, PGIS, IP and PPARδ protein levels after stromal cells were treated with AA for 6 hr. (F) Western blot analysis on effects of NS398 (COX-2 inhibitor) on AA induction of COX2, PGES, PGIS, IP and PPARδ protein levels after stromal cells were treated with AA for 48 hr in the absence or presence of NS398. (G) QPCR analysis of Inhba mRNA level after stromal cells were treated with AA for 72 hr. (H) QPCR analysis of on effects of NS398 on AA induction of Prl8a2 mRNA level after stromal cells were treated with AA for 72 hr. (I) QPCR analysis of on effects of GSK0660 on AA induction of Prl8a2 mRNA level after stromal cells were treated with AA for 72 hr. (J) Western blot analysis on effects of AA on ACTIVIN A protein level after stromal cells were treated with AA for 24 hr. (K) QPCR analysis on effects of SB431542 (ACTIVIN A inhibitor) on AA induction of Prl8a2 mRNA levels. (L) QPCR analysis on the interference efficiency of Inhba. (M) QPCR analysis on effects of Inhba siRNAs on AA induction on Inhba mRNA level after stromal cells were treated with AA for 72 hr. Data were presented as means ± SD from at least three biological replicates. *, p<0.05; **, p<0.01; ***, p<0.001. CYC D3: CYCLIN D3; ACT-A: ACTIVIN A.
-
Figure 4—source data 1
Raw data of all western blots from Figure 4.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig4-data1-v2.zip
-
Figure 4—source data 2
Complete and uncropped membranes of all western blots from Figure 4.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig4-data2-v2.zip
Blastocyst-derived TNF promotes CPLA2α phosphorylation and AA secretion
We just showed that AA could initiate fibroblast activation and induce decidualization. When we examined the concentration of AA in the uterine luminal fluid, the luminal concentration of AA at D4.5 was significantly higher than that on D3 and D4, indicating AA secretion from the uterus just after embryo implantation (Figure 5A).
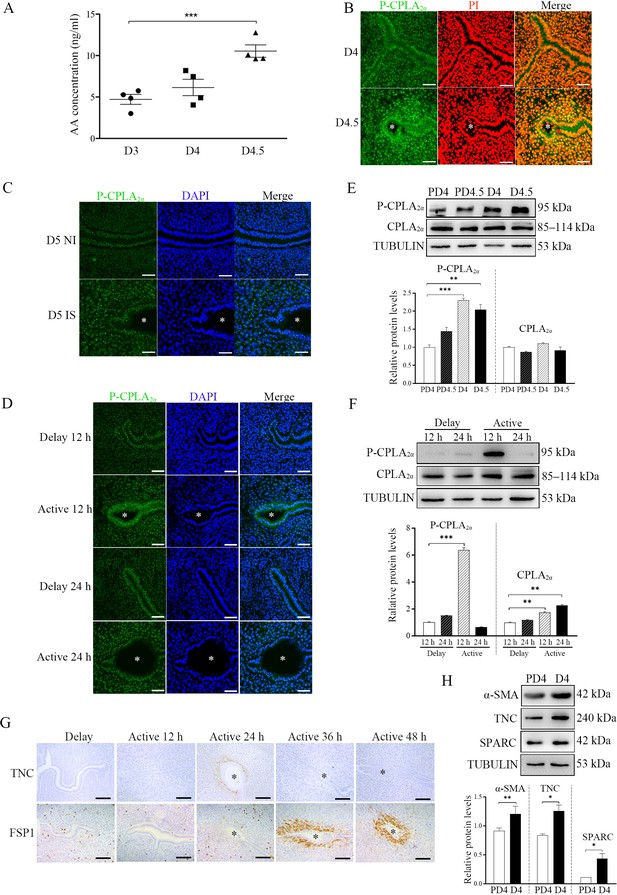
The involvement of blastocysts in fibroblast activation during early pregnancy.
(A) AA concentration in uterine luminal fluid flushed on D3 (n=20 mice), D4 (n=20 mice), and D4.5 (n=20 mice) of pregnancy. (B) P-CPLA2α immunofluorescence in mouse uteri on D4 (n=6) and D4.5 (n=6). * Embryo. Scale bar = 50 μm. (C) P-CPLA2α immunofluorescence in mouse uteri at implantation sites and inter-implantation sites on D5 (n=6 mice). * Embryo. NI, inter-implantation site; IS, implantation site. Scale bar = 50 μm. (D) p-CPLA2α immunofluorescence of in mouse uteri 12 and 24 h after delayed implantation was activated by estrogen treatment, respectively (n=4 mice). * Embryo. Scale bar = 0 μm. (E) Western blot analysis of CPLA2α and P-CPLA2α protein levels in mouse uteri on D4, D4.5 PD4 and PD4.5 (n=4 mice), respectively. (F) Western blot analysis of CPLA2α and P-CPLA2α protein levels in mouse uteri 12 and 24 hr after delayed implantation was activated by estrogen treatment (n=4 mice). (G) Immunostaining of TNC and S100A4 in mouse uteri 12, 24, 36, and 48 hr after delayed implantation was activated by estrogen treatment (n=4 mice). * Embryo. (H) Western blot analysis α-SMA, TNC, and SPARC protein levels in mouse uteri on D4 and PD4 (n=4 mice). *, p<0.05; **, p<0.01; ***, p<0.001.
-
Figure 5—source data 1
Raw data of all western blots from Figure 5.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig5-data1-v2.zip
-
Figure 5—source data 2
Complete and uncropped membranes of all western blots from Figure 5.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig5-data2-v2.zip
Immunofluorescence also showed that the P-CPLA2α level in luminal epithelium in D4.5 was obviously stronger than that on D4 (Figure 5B). Compare with inter-implantation sites, P-CPLA2α immunofluorescence at implantation site on D5 was also stronger (Figure 5C). Compared with delayed implantation, P-CPLA2α immunofluorescence in the luminal epithelium was stronger 12 hr after estrogen activation (Figure 5D). Western blot also confirmed that P-CPLA2α levels on D4 and D4.5 were significantly higher than PD4 and PD4.5 (Figure 5E). Compared with delayed implantation, P-CPLA2α protein levels were also higher 12 hr after estrogen activation (Figure 5F). These results suggested that embryos should be involved in CPLA2α phosphorylation.
Furthermore, we examined the markers of fibroblast activation. Compared with delayed implantation, the immunostaining levels of both TNC and S100A4 at implantation sites after estrogen activation were stronger (Figure 5G). Western blot also indicated that the protein levels of the markers of fibroblast activation (αSMA by 1.32-folds, TNC by 1.51-folds, and SPARC by 4.0-folds) on D4 were stronger than that on PD4 (Figure 5H). This results strongly suggest that embryos were tightly involved in fibroblast activation.
S100A9, HB-EGF and TNF are previously shown to be secreted by blastocysts (He et al., 2019; Haller et al., 2019; Hamatani et al., 2004). When S100A9-soaked blue beads were transferred into PD4 uterine lumen, S100A4 had no obvious change, but TNC immunostaining was increased slightly (Figure 6A). When HB-EGF-soaked beads were transferred, S100A4 immunostaining was slightly increased, but TNC immunostaining was increased obviously (Figure 6B). However, TNF-soaked beads obviously stimulated the immunostaining levels of both S100A4 and TNC (Figure 6C).
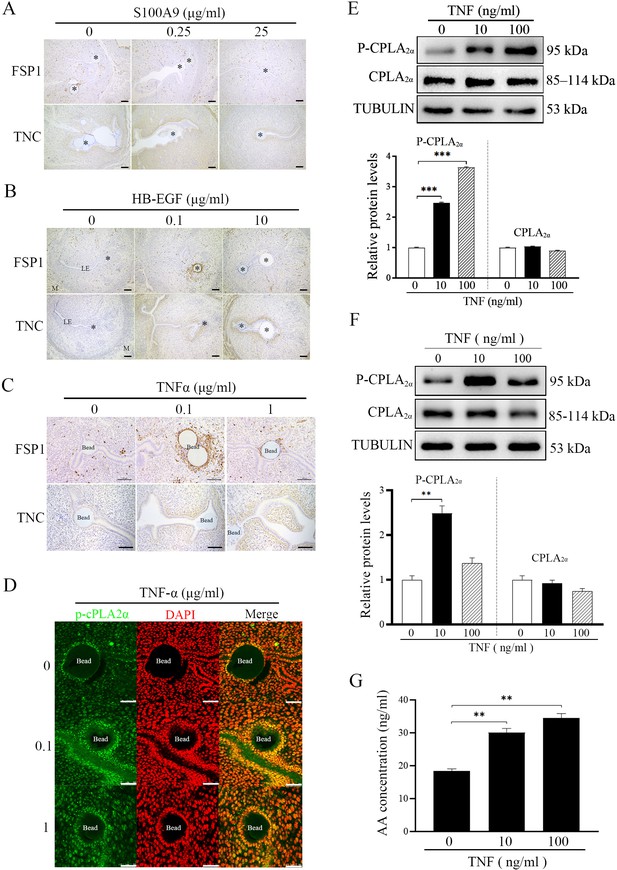
Effects of TNF on CPLA2α phosphorylation and AA secretion.
(A) Immunostaining of TNC and S100A4 in mouse uteri after S100A9-soaked blue beads were transferred into uterine lumen of PD4 mice for 24 hr (n=6 mice). (B) Immunostaining of TNC and S100A4 in mouse uteri after HB-EGF-soaked blue beads were transferred into uterine lumen of PD4 mice for 24 hr (n=6 mice). (C) Immunostaining of TNC and S100A4 in mouse uteri after TNF-soaked blue beads were transferred into uterine lumen of PD4 mice for 24 hr (n=6 mice).* Bead; LE, luminal epithelium; M, muscular layer; St, stroma. Scale bar = 100 μm. (D) P-CPLA2α immunofluorescence in mouse uteri after 0.1 and 1 μg/ml TNF-soaked blue beads were transferred into PD4 uterine lumen (n=3 mice). Scale bar = 50 μm. (E) Western blot analysis of CPLA2α and P-CPLA2α protein levels after cultured epithelial cells were treated with TNF for 3 hr. (F) Western blot analysis of CPLA2α and P-CPLA2α protein levels after cultured epithelial organoids were treated with TNF for 3 hr. (G) ELISA analysis of AA concentration in the cultured medium after cultured epithelial organoids were treated with TNF for 6 hr. *, p<0.05; **, p<0.01; ***, p<0.001.
-
Figure 6—source data 1
Raw data of all western blots from Figure 6.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig6-data1-v2.zip
-
Figure 6—source data 2
Complete and uncropped membranes of all western blots from Figure 6.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig6-data2-v2.zip
After TNF-soaked beads were transferred into PD4 uterine lumen, P-CPLA2α immunofluorescence at luminal epithelium was obviously increased (Figure 6D). When the epithelial cells isolated PD4 uterus were treated with TNF, western blot showed that the protein level of P-CPLA2α was significantly upregulated 2.31-folds (Figure 6E). To confirm the results from in vitro cultured epithelial cells, mouse uterine epithelial organoids was treated with TNF. We found that the expression of P-CPLA2α was up-regulated 2.49-folds by TNF treatment in mouse uterine epithelial organoids (Figure 6F). AA concentration in the cultured medium of epithelial orgoids was also up-regulated 1.88-folds by TNF treatment (Figure 6G). These results indicated that embryo-derived TNF was able to promote the phosphorylation of CPLA2α in luminal epithelium for liberating AA into uterine stroma.
Effects of fibroblast activation on TNF-AA-PGI2 pathway under human in vitro decidualization
After we showed that fibroblast activation was involved in mouse decidualization, we wondered whether fibroblast activation participated in human decidualization. Compared with control group, the protein levels of α-SMA, TNC and SPARC were significantly increased under human in vitro decidualization (1.30, 5.12, and 2.29 folds; Figure 7A). Previous studied also indicated that human blastocysts could synthesize and secret TNF (He et al., 2019; Lv et al., 2022a). When human uterine epithelial Ishikawa cells were co-cultured with human 4003 stromal cells, TNF treatment significantly increased the protein level of P-CPLA2α by 1.59-folds in epithelial Ishikawa cells (Figure 7B), and the protein levels of TNC, αSMA, SPARC in 4003 stromal cells (2.71, 2.44, and 1.96 folds) (Figure 7C). When human stromal cells were treated with AA, the protein levels of TNC, αSMA, and SPARC were obviously stimulated (Figure 7D). Meanwhile, treatment with AA also significantly upregulated the protein levels of COX-2, PGIS, and PPARδ (1.28, 4.69, and 1.60 folds), but had no obvious effects on PGES and IP protein level (Figure 7E). After 4003 stromal cells were treated with either PGI2 analogs ILOPROST or PPARδ agonists GW501516, the protein levels of TNC, αSMA, and SPARC were significantly up-regulated compared with control group (2.18, 1.57, and 1.38 or 1.79, 2.86, and 3.73 folds; Figure 7F and G). These results suggested that AA should promote fibroblast activation through PGI2–PPARδ pathway during human decidualization. Treatment with AA also stimulated the mRNA expression of INHBA (3.4-folds; ncoded for human ACTIVIN A) (Figure 7H), which was significantly abrogated by COX-2 inhibitor NS398 (1.88-folds; Figure 7I). Overall, these results indicated that fibroblast activation was also involved in human decidualization in a similar mechanism as in mice.
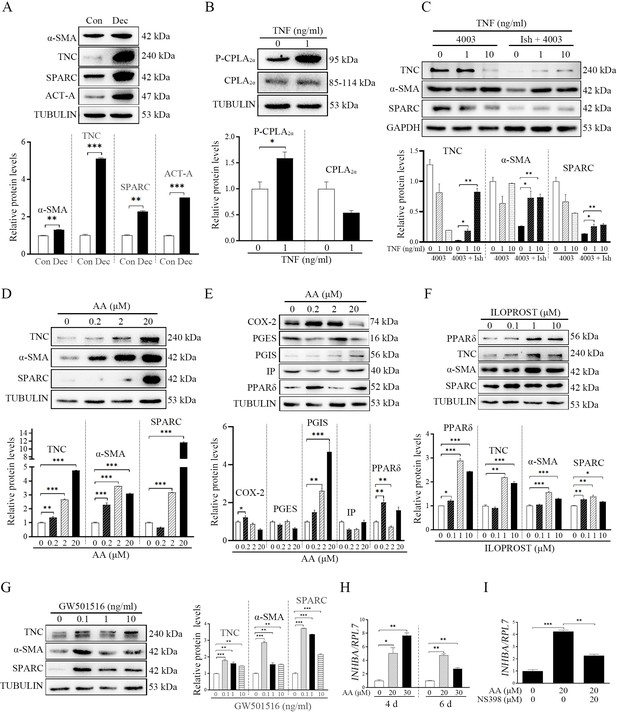
TNF regulation of fibroblast activation through AA-PGI-ACTIVIN A pathway in human endometrium.
(A) Western blot analysis of α-SMA, TNC, SPARC, and ACTIVIN A protein levels after human stromal cells were induced for decidualization for 24 hr. ACT-A: ACTIVIN A. (B) Western blot analysis of CPLA2α and P-CPLA2α protein levels in Ishikawa cells after the co-culture of Ishikawa cells and 4003 cells were treated with TNF for 3 hr. (C) Western blot analysis of TNC, α-SMA, and SPARC protein levels in stromal 4003 cells after the co-culture of Ishikawa cells and 4003 cells were treated with TNF for 3 hr. Ish: Ishikawa. (D) Western blot analysis of TNC, α-SMA and SPARC protein levels after stromal 4003 cells were treated with AA for 6 hr. (E) Western blot analysis of COX-2, PGES, PGIS, and PPARδ protein levels after stromal 4003 cells were treated with AA for 3 hr. (F) Western blot analysis of PPARδ, TNC, α-SMA and SPARC protein levels after stromal 4003 cells were treated with ILOPROST for 12 hr. (G) Western blot analysis of TNC, α-SMA, and SPARC protein levels after stromal cells 4003 cells were treated with GW501516 for 6 hr. (H) QPCR analysis of INHBA mRNA levels after stromal 4003 cells were treated with AA. (I) QPCR analysis on effects of NS398 on AA stimulation of INHBA mRNA levels after stromal 4003 cells were treated with AA. *, p<0.05; **, p<0.01; ***, p<0.001.
-
Figure 7—source data 1
Raw data of all western blots from Figure 7.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig7-data1-v2.zip
-
Figure 7—source data 2
Complete and uncropped membranes of all western blots from Figure 7.
- https://cdn.elifesciences.org/articles/82970/elife-82970-fig7-data2-v2.zip
AA-PGI2 pathway contributes to human decidualization
Insulin growth factor binding protein 1(IGFBP1) and prolactin (PRL) are recognized markers for human in vitro decidualization (Saleh et al., 2011). When human stromal cells were treated with AA, both IGFBP1 and PRL were significantly increased by 4.80 and 3.87-folds, respectively (Figure 8A). Under human in vitro decidualization (MPA +cAMP 100 μM), either ILOPROST or GW501516 could significantly promote the mRNA expression of IGFBP1 and PRL (212.20 and 42.30 folds, or 4363.93 and 36.39 folds) (Figure 8B and C). The induction of AA on IGFBP1 and PRL mRNA expression was significantly abrogated by either COX-2 inhibitor NS398 or PPARδ antagonist GSK0660 (1.85 and 4.63 folds; Figure 8D). Under human in vitro decidualization (MPA +cAMP 500 μM), the mRNA levels of IGFBP1 and PRL were also significantly up-regulated by ACTIVIN A treatment (6.62 and 1.61 folds; Figure 8E). AA-induced increase of IGFBP1 and PRL expression was abrogated by INHBA knockdown (Figure 8F and G). Taken together, these results suggested that epithelium-derived AA could promote fibroblast activation through COX-2-PGI2-PPARδ pathway and induce decidualization via ACTIVIN A during human decidualization, which was also consistent with the results in mice.
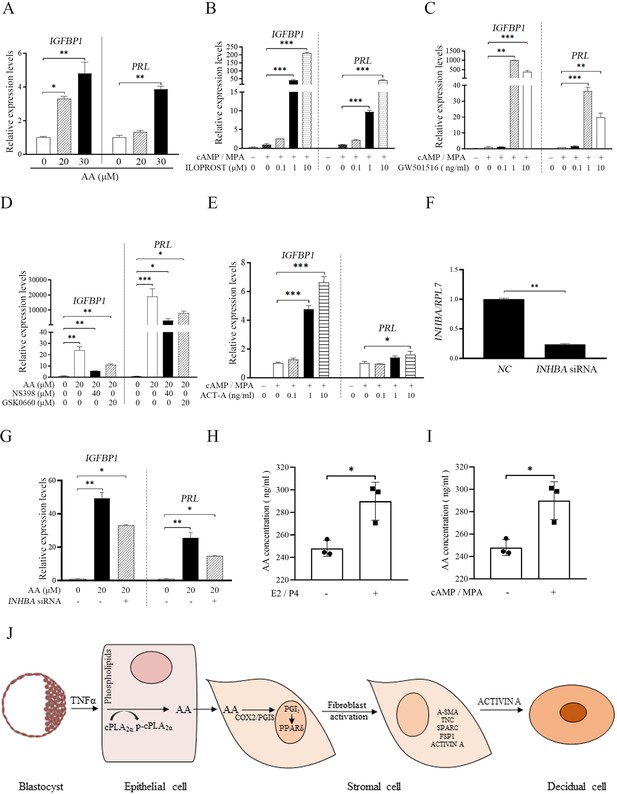
AA-PGI2-PPARδ-ACTIVIN A regulation on human decidualization.
(A) QPCR analysis of IGFBP1 and PRL mRNA levels after stromal 4003 cells were treated with AA. (B) QPCR analysis of IGFBP1 and PRL mRNA levels after stromal 4003 cells were treated with ILOPROST under in vitro decidualization for 4 days. (C) QPCR analysis of IGFBP1 and PRL mRNA levels after stromal 4003 cells were treated with GW501516 under in vitro decidualization for 4 days. (D) QPCR analysis on effects of NS398 or GSK0660 on AA induction of IGFBP1 and PRL mRNA levels after stromal 4003 cells were treated with AA for 4 days. (E) QPCR analysis of IGFBP1 and PRL mRNA levels after stromal 4003 cells were treated with ACTIVIN A for 2 days under in vitro decidualization. ACT-A: ACTIVIN A. (F) QPCR analysis on the interference efficiency of INHBA. (G) QPCR analysis on effects of INHBA siRNAs on AA induction of IGFBP1 and PRL mRNA levels in stromal 4003 cells for 4 days. (H) AA concentration in the cultured medium after mouse stromal cells were induced for in vitro decidualization with estrogen (E2) and progesterone (P4) for 24 h. (I) AA concentration in the cultured medium after human endometrial stromal 4003 cells were induced for in vitro decidualization with MPA and cAMP for 4 days. (J) The model of fibroblast activation action in the blastocyst-endometrial interaction. *, p<0.05; **, p<0.01; ***, p<0.001.
Discussion
Our study shows that embryo-derived TNF promotes the phosphorylation of CPLA2α and the release of AA from luminal epithelium to induce fibroblast activation and decidualization through ACTIVIN A. We also showed that the pathway underlying fibroblast activation was conserved in mice and humans.
Fibroblast activation could be identified by many markers, including α-SMA, TNC, POSTN, NG-2, PDGF receptor-a/b, S100A4and FAP. These fibroblast markers expressed alone or in combination and could be used to identify distinct subpopulations following fibroblast activation (Prime et al., 2017; Grinnell, 1994; Oliver et al., 1999). Fibroblast activation plays an important role under many physiological or pathological conditions, such as cancer, injury repair and fibrosis. However, the regulation and function of fibroblast activation during decidualization remain unknown. In our study, the regulation and function of fibroblast activation was confirmed by multiple approaches during mouse and human decidualization. A study from single-cell analysis of human decidual cells also suggests the presence of fibroblast activation during human in vitro decidualization (Stadtmauer and Wagner, 2022). Based on fibroblast activation markers, we confirmed the existence of fibroblast activation during early pregnancy. Fibroblasts are generally in a dormant and quiescent state in tissues, and are only activated when stimulated (Pakshir et al., 2020). In a previous study, mPGES1-derived PGE2 supports the early inflammatory phase of wound healing and may stimulate subsequent fibroblast activation early after damage (Stratton and Shiwen, 2010). Although PGE2 has been shown to be essential for mouse decidualization (Holmes and Gordashko, 1980; Baskar et al., 1981), PGE2 was ineffective in activating fibroblast activation in our study. Additionally, PGE2 has no obvious effect on Prl8a2 mRNA level under mouse in vitro decidualization. Our study indicated that AA from luminal epithelium was able to induce fibroblast activation via PGI2-PPARδ pathway. Previous studies indicated that AA could promote mouse decidualization (Tessier-Prigent et al., 1999; Handwerger et al., 1981). PGI2 has been identified to be essential for mouse decidualization via PPARδ (Lim et al., 1999). It is shown that multiple factors are able to induce in vitro decidualization (Gellersen and Brosens, 2014). In our study, AA-PGI2 pathway is involved in stimulating decidualization. previous studies indicate that PGI2 can also activate a cAMP signaling pathway (Beck et al., 2014; Fetalvero et al., 2008). It is possible that AA may also act on decidualization through cAMP. Our study also confirmed AA secretion during either mouse or human in vitro decidualization, which may have positive feedback during in vitro decidualization. In our study, AA concentration in uterine lumen was significantly increased during embryo attachment. AA, PGI2 or PPARδ agonist was able to induce fibroblast activation.
Activated fibroblasts play key roles in the injury response, tumorigenesis, fibrosis, and inflammation through secreting different factors in different physiological or pathological processes (Avery et al., 2018). Myofibroblast marker α-SMA has been shown to be one of the important markers of early decidualization in primates (Kim et al., 1999), and sterile inflammatory secretion of products such as ATP and uric acid after injury has been shown to stimulate fibroblast activation (Dolmatova et al., 2012; Bao et al., 2018) and uterine decidualization (Gu et al., 2020; Zhu et al., 2021), so we speculate that fibroblast activation is strongly correlated with decidualization. Because α-SMA, TNC, SPARC, S100A4, and ACTIVIN A were all identified during fibroblast activation, we examined the role of each of these markers during mouse decidualization. We found that only ACTIVIN A was able to induce mouse in vitro decidualization. The function of ACTIVIN A during human decidualization was also confirmed in our study. ACTIVIN A has been shown to be important for human decidualization (Zhao et al., 2018). ACTIVIN A, its functional receptors, and binding proteins, are abundant in human endometrium (He et al., 1999). Our study indicated that AA-PGI2-PPARδ axis stimulated fibroblast activation and induced decidualization through ACTIVIN A.
The adequate molecular interaction between the endometrium and the blastocyst is critical for successful implantation and decidualization (Massimiani et al., 2019; Latifi et al., 2018). In our study, there was a big difference of both markers of fibroblast activation and P-CPLA2α between pregnancy and pseudopregnancy, and between delayed and activated implantation, strongly suggesting the involvement of embryos in these processes. Although S100A9, HB-EGF, and TNF are previously shown to be secreted by blastocysts (He et al., 2019; Haller et al., 2019; Hamatani et al., 2004), TNF was the only one to stimulate both S100A4 and TNC, and to phosphorylate CPLA2α in our study. CPLA2α is a major provider of AA. Pla2g4a–/–mice results in deferred implantation and deranged gestational development (Song et al., 2002). We also showed that AA concentration in luminal fluid was significantly increased in day 4 evening when the embryos just implanted. TNF is present in the reactivated blastocyst and human blastocyst (He et al., 2019; Lv et al., 2022a), and may play a critical role during embryo implantation (You et al., 2021). In our study, we confirmed that TNF stimulated the phosphorylation of CPLA2α and AA release from luminal epithelium. A proper interaction between embryo attachment and decidualization is critical for successful pregnancy.
Activated fibroblasts play an important role in many physiological and pathologic processes. Excessive fibroblast activation can lead to fibrosis (Truffi et al., 2020; Giusti et al., 2022; Yeo et al., 2018). Fibroblast activation may be present and balanced during normal pregnancy without ultimately leading to fibrosis or other diseases. S100A4 is hypomethylated and overexpressed in grade 3 endometrioid tumors compared with benign endometrium (Xie et al., 2007). Genetic and proteomic analysis of surgical specimens from 14 patients with uterine leiomyoma showed that TNC is significantly upregulated in patient samples (Jamaluddin et al., 2019). Fibrosis mostly occurs in pathological conditions. Intrauterine adhesions (IUA), also known as Asherman’s syndrome, are caused by endometrial fibrosis as a result of injury to the uterus’s basal lining, resulting in partial or total adhesions in the uterine cavity (Healy et al., 2016). IUA can interfere with the embryo’s implantation and development, resulting in decreased or even full loss of intrauterine volume, female infertility, and recurrent miscarriages (Leung et al., 2021). The thin endometrial model exhibits a higher degree of fibrosis than normal controls, which is thought to be a crucial component in embryo implantation failure (Zhang et al., 2022). The key question is why fibrosis doesn’t occur during normal early pregnancy? Both activins and inhibits are expressed in pregnant uterus (Ni and Li, 2017). In our study, Fst was significantly up-regulated under in vitro decidualization although ACTIVIN A was able to stimulate in vitro decidualization. FST is a secreted glycoprotein and can neutralize the profibrotic and proinflammatory actions of ACTIVINS. FST has a strong antifibrotic effect in various organs (Aoki et al., 2005; Patella et al., 2006). Furthermore, FST is shown to be critical for mouse decidualization (Fullerton et al., 2017). Additionally, AA was able to induce fibroblast activation and promote decidualization in our study. A recent study showed that 11,12-epoxyeicosatrienoic acid, a metabolite of AA can alleviate pulmonary fibrosis (Kim et al., 2021). It is possible that a physiological level of fibroblast activation is beneficial for decidualization and the long-lasting fibroblast activation could be balanced by certain molecules, like FST or AA metabolite.
Conclusion
In this study, we identified that embryos-derived TNF is able to phosphorylate CPLA2α for releasing AA from luminal epithelium. AA can physiologically induce fibroblast activation and promote decidualization via PGI2-PPARδ-ACTIVIN A axis. This regulatory mechanism is also conserved in mice and humans. Overall, this study should shed a light on the novel mechanism underlying decidualization (Figure 8J).
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-SPARC (rabbit monoclonal) | Cells Signaling Technology | Cat. #: 8725 S RRID: AB_2617210 | 1:200(IF) 1:1000(WB) |
| Antibody | Anti-Phospho-cPLA2 (rabbit polyclonal) | Cells Signaling Technology | Cat. #: 2831 S RRID: AB_2164445 | 1:100(IF) 1:1000(WB) |
| Antibody | Anti- Tenascin C (rabbit monoclonal) | Abcam | Cat. #: ab108930 RRID: AB_10865908 | 1:200(IHC) 1:1000(WB) |
| Antibody | Anti- S100A4 (rabbit monoclonal) | Cells Signaling Technology | Cat. #: 13018 S RRID: AB_10865908 | 1:200(IHC) 1:1000(WB) |
| Antibody | Anti- CPLA2α(rabbit polyclonal) | Santa Cruz Biotechnology | Cat. #: SC-438 RRID: AB_2164442 | 1:1000 |
| Antibody | Anti-α-SMA (rabbit monoclonal) | Cells Signaling Technology | Cat. #: 19,245T RRID: AB_476977 | 1:1000 |
| Antibody | Anti- COX-2 (rabbit monoclonal) | Cells Signaling Technology | Cat. #: 12,282T RRID: AB_1149420 | 1:1000 |
| Antibody | Anti- PGIS (rabbit polyclonal) | Cayman Chemical | Cat. #: 100023 RRID: AB_10816097 | 1:1000 |
| Antibody | Anti- PPAR delta (rabbit monoclonal) | Abcam | Cat. #: ab178866 RRID: AB_2165907 | 1:1000 |
| Antibody | Anti- BMP2 (rabbit polyclonal) | Abclonal | Cat. #: A0231 RRID: AB_2313822 | 1:1000 |
| Antibody | Anti- WNT4 (mouse monoclonal) | Santa Cruz Biotechnology | Cat. #: sc-376279 RRID: AB_787604 | 1:1000 |
| Antibody | Anti- E2F8 (rabbit polyclonal) | Abclonal | Cat. #: sc-376279 RRID: AB_10639701 | 1:1000 |
| Antibody | Anti- CYCLIN D3 (mouse monoclonal) | Cells Signaling Technology | Cat. #: 2936T RRID: AB_397675 | 1:1000 |
| Antibody | Anti-α- TUBULIN (mouse monoclonal) | Cells Signaling Technology | Cat. #: 2144 S RRID: AB_2716367 | 1:1000 |
| Antibody | Anti- GAPDH (mouse monoclonal) | Santa Cruz Biotechnology | Cat. #: sc-32233 RRID: AB_307275 | 1:1000 |
| Antibody | Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, HRP (Goat polyclonal) | inventrogen | Cat. #: G21234 RRID: AB_2534776 | 1:5000 |
| Antibody | Alexa Fluor 488-AffiniPure Goat Anti-Rabbit IgG (H+L) (Goat polyclonal) | Jackson ImmunoResearch | Cat. #: 111-545-144 RRID:AB_2338046 | 1:200 |
| Chemical compound, drug | Recombinant Mouse TNF-alpha (aa 80–235) Protein | R&D systems | Cat. #: 410-MT-010 | 1–100 ng/ml(recombinant protein) |
| Chemical compound, drug | TNC | R&D systems | Cat. #: 410-MT-010 | 5–500 ng/ml(recombinant protein) |
| Chemical compound, drug | S100A4 | MedChemExpress | Cat. #: HY-P71084 | 50 and 500 ng/ml(recombinant protein) |
| Chemical compound, drug | SPARC | MedChemExpress | Cat. #: HY-P71086 | 1 and 10 μM (recombinant protein) |
| Chemical compound, drug | AA | Sigma | Cat. #: A3611 | 0.2 and 20 μM (recombinant protein) |
| Chemical compound, drug | ILOPROST | MedChemExpress | Cat. #: HY-A0096 | 0.1 and 10 μM (recombinant protein) |
| Chemical compound, drug | SELEXIPAG | MedChemExpress | Cat. #: HY-14870 | 0.1 and 10 μM (recombinant protein) |
| Chemical compound, drug | GW501516 | Cayman Chemical | Cat. #: 317318-70-0 | 0.1 and 10 μM (recombinant protein) |
| Chemical compound, drug | NS 398 | Selleck | Cat. #: S8433 | 20 and 40 μM(recombinant protein) |
| Chemical compound, drug | GSK0660 | Selleck | Cat. #: S5817 | 40 μMμM(recombinant protein) |
| Chemical compound, drug | ACTIVIN A | MedChemExpress | Cat. #: HY-P70311 | 1–100 ng/ml(recombinant protein) |
| Cell line (Homo-sapiens) | T HESCs | ATCC | CRL-4003 | |
| Cell line (Homo-sapiens) | Ishikawa | Chinese Academy of Science | CRL-2923 | |
| Commercial assay or kit | AA ELISA kit | Elabscience | Cat. #: E-EL-0051c | |
| Other | PI stain | Sigma | Cat. #: P4170 | 5 μg/ml |
| Other | DAPI stain | Sigma | Cat. #: D9542 | 20 μg/ml |
| Software, algorithm | SPSS | SPSS | RRID:SCR_002865 | |
| Software, algorithm | Image J | ImageJ (http://imagej.nih.gov/ij/) | RRID:SCR_003070 | |
| Software, algorithm | GraphPad Prism software | GraphPad Prism (https://graphpad.com) | RRID:SCR_015807 | Version 8.0.0 |
Animals and treatments
Request a detailed protocolMature CD1 mice (Hsd: ICR mouse, 6 weeks of age) were purchased from Hunan Slaike Jingda Laboratory Animal Co., LTD and maintained in specific pathogen free (SPF) environment and controlled photoperiod (light for 12 hr and darkness for 12 hr). All animal protocols were approved by the Animal Care and Use Committee of South China Agricultural University (No. 2021f085). In order to induce pregnancy or pseudopregnancy, female mice aged 8–10 weeks were mated with male mice of reproductive age or vasectomized mice (vaginal plug day for day 1). To confirmed the pregnancy of female mice, embryos were flushed from fallopian tubes or uteri from days 1 to 4. On days 4 midnight (D4.5), 5 (D5) and 5 midnight (D5.5), implantation sites were identified by intravenous injection of 0.1 ml of 1% Chicago blue dye (Sigma-Aldrich, St. Louis, MO) dissolved in saline. Uteri were also collected from pseudopregnant mice on day 4 (PD4) , D4.5 , D5, and D5.5 .
On day 4 of pregnancy (0800–0900 h) (D4), pregnant mice were ovariectomized to induce delayed implantation. From days 5 to 7, progesterone was injected daily (1 mg/0.1 ml sesame oil/mice, Sigma-Aldrich) to maintain delayed implantation. Estradiol-17β (1 μg/ml sesame oil/mouse, MCE) (Diao et al., 2008) was subcutaneously injected on day 7 to activate embryo implantation. Delayed implantation was confirmed by flushing the blastocyst from the uterine horn. The implantation site of the activated uterus was determined by intravenous injection of Chicago blue dye.
Transfer of TNF-soaked beads
Request a detailed protocolAffi-Gel Blue Gel Beads (Bio-Rad # 1537302) with blastocyst size were incubated with TNF (0.1% BSA/PBS 410-MT-010, R&D systems, Minnesota, USA) 37 °C for 4 hr. After washed with embryo culture medium M2 (M7167, Sigma Aldrich, St. Louis, MO) (0.1% BSA) for three times, TNF-soaked beads (15 beads/horn) were transplanted into the uterine horns of PD4 mice (n=6 mice). Beads incubated in 0.1% BSA/PBS were used as control group (n=6 mice). Blue bands with beads were identified by injecting Chicago blue into the tail intravenous injection of Chicago blue to observe the implantation site 3 and 24 hr after transplantation, respectively.
Immunofluorescence
Request a detailed protocolImmunofluorescence was performed as described previously (Li et al., 2020; Zheng et al., 2020). Briefly, frozen sections (10 μm) were fixed in 4% paraformaldehyde (158127, Sigma Aldrich) for 10 min. Frozen or paraffin sections were blocked with 10% horse serum for 1 hr at 37 °C and incubated overnight with appropriate dilutions of primary antibodies at 4 °C. The primary antibodies used in this study included anti-SPARC (1:200, 8725 S, Cell Signaling Technology, Danvers, MA) and anti-P-CPLA2α (1:100, 2831 S, Cell Signaling Technology). After washing in PBS, sections were incubated with secondary antibody (2.5 μg/ml, G21234, Invitrogen, Carlsbad, CA) for 40 min, counterstained with 4, 6-diamidino-2-phenylindole dihydrochloride (20 μg/ml, DAPI, D9542, Sigma-Aldrich) or propidium iodide (5 μg/ml, PI, P4170, Sigma-Aldrich) and were mounted with ProLong Diamond Antifade Mountant (Thermo Fisher, Waltham, MA). The pictures were captured by laser scanning confocal microscopy (Leica, Germany). Each experiment was repeated at least three times.
Immunohistochemistry
Request a detailed protocolImmunohistochemistry was performed as described previously (Fu et al., 2019). In short, paraffin sections (5 μm) were deparaffined, rehydrated, and antigen retrieved by boiling in 10 mM citrate buffer for 10 min. Endogenous horseradish peroxidase (HRP) activity was inhibited with 3% H2O2 solution in methanol. After washing for three times with PBS, the sections were incubated at 37℃ for 1 hr in 10% horse serum for blocking, incubated overnight in each primary antibody at 4 °C. The primary antibodies used in this study included anti-TNC (1:200, ab108930, Abcam, Cambridge, UK) and anti-S100A4 (1:200, 13018 S, Cell Signaling Technology). After washing, the sections were incubated with biotinylated rabbit anti-goat IgG antibody (1:200, Zhongshan Golden Bridge, Beijing, China) and streptavidin-HRP complex (1:200, Zhongshan Golden Bridge). According to the manufacturer’s protocol, the positive signals were visualized using DAB Horseradish Peroxidase Color Development Kit (Zhongshan Golden Bridge). The nuclei were counter-stained with hematoxylin. Each experiment was repeated at least three times.
siRNA transfection
Request a detailed protocolThe siRNAs for mouse Activin A were designed and synthesized by Ribobio Co., Ltd. (Guangzhou, China). Following manufacturer’s protocol, mouse endometrial stromal cells or human endometrial stromal cell 4003 (ATCC, CRL-4003) (American Type Culture Collection) were transfected with each Inhba siRNA using Lipofectamine 2000 Transfection Reagent (Invitrogen, Grand Island, NY) for 6 hr and 12 hr, respectively. The siRNA sequences were listed in Table 1. Each experiment was repeated at least three times.
Primers and siRNA sequences used in this study.
| Gene | Species | Sequence (5’–3’) | Application | Accession Number | Product size |
|---|---|---|---|---|---|
| Rpl7 | Mouse | GCAGATGTACCGCACTGAGATTC ACCTTTGGGCTTACTCCATTGATA | RT-QPCR | NM_011291.5 | 129 bp |
| Prl8a2 | Mouse | AGCCAGAAATCACTGCCACT TGATCCATGCACCCATAAAA | RT-QPCR | NM_010088 | 119 bp |
| Inaba | Mouse | CCAGTCTAGTGCTTCTGGGC GATGAGGGTGGTCTTCGGAC | RT-QPCR | NM_008380.2 | 156 bp |
| RPL7 | Human | CTGCTGTGCCAGAAACCCTT TCTTGCCATCCTCGCCAT | RT-QPCR | NM_000971 | 194 bp |
| IGFBP1 | Human | CCAAACTGCAACAAGAATG GTAGACGCACCAGCAGAG | RT-QPCR | NM_001013029 | 87 bp |
| PRL | Human | AAGCTGTAGAGATTGAGGAGCAAA TCAGGATGAACCTGGCTGACTA | RT-QPCR | NM_000948 | 76 bp |
| INHBA | Human | TCATCACGTTTGCCGAGTCA TGTTGGCCTTGGGGACTTTT | RT-QPCR | NM_002192 | 129 bp |
| NC | - | CTCCGAACGTGTCACGT | siRNA | ||
| Activin a | Mouse | GAACAGTCACATAGACCTT | siRNA | ||
| ACTIVIN A | Human | CCAUGUCCAUGUUGUACUATT | siRNA |
AA assay
Request a detailed protocolMouse uteri on D3, D4, and D4.5 (n=5 mice per day) were flushed with 0.4 ml of normal saline per mouse. The flushing solution from 5 mice was combined and dried in a vacuum freeze drier at –80 °C for 24 hr. The dried product was dissolved in 400 μl of the working buffer for AA measurement. After mouse epithelial organoids were treated with TNF (10 and 100 ng/ml) for 6 hr (n=3), the cultured medium was collected for AA measurement. AA amount in the uterine fluid or cultured medium was measured with AA ELISA kit according to the manufacturer’s instruction (Elabscience, E-EL-0051c, Wuhan, China). This kit’s sensitivity is greater than 0.94 ng/ml. In brief, 50 μl of each sample was incubated at 37 °C for 45 min with 50 μl of biotinylated antibody working solution, 100 μL of HRP enzyme conjugate working solution for 30 min, and 90 μl of substrate solution for 15 min before being stopped with 50 μl of substrate solution. The solution was immediately read at 450 nm with a Biotek microplate reader (ELX808). Absorbance values for AA standards were calculated in the same way. The AA standard curve was used to calculate the concentrations of AA.
Isolation and treatment of mouse uterine luminal epithelial cells
Request a detailed protocolUterine luminal epithelial cells were isolated as previously described (Nallasamy et al., 2012). The uteri from the estrous mice or PD4 were cut longitudinally, washed in HBSS, incubated in 0.2% (W/V) trypsin (0458, Amresco, Cleveland, USA) and 6 mg/ml dispase (Roche Applied Science,4942078001, Basel, Switzerland) in 4.3 ml HBSS for 1.5 hr at 4℃, 30 min at room temperature, and 10 min at 37℃. After rinsing in HBSS, the epithelial cells were precipitated in 5% BSA in HBSS for 7 min. After the collected epithelial cells were cultured in DMEM/F12 (D2906, Sigma-Aldrich) with 10% heat-inactivated fetal bovine serum (FBS) in a culture plate for 30 min, the unattached epithelial cells were transferred into new culture plates precoated with ECM (1:100, E0282, Sigma-Aldrich) and cultured at 37 °C for 1 hr. Luminal epithelial cells were treated with TNF (10 and 100 ng/ml, 410-MT-010, R&D systems) in DMEM/F12 with 2% charcoal-treated FBS (cFBS, Biological Industries, Cromwell, CT).
Culture and treatment of mouse uterine luminal epithelial organoids
Request a detailed protocolUterine luminal epithelial organoids were isolated and cultured as previously described (Turco et al., 2017; Boretto et al., 2017; Luddi et al., 2020). Mouse uteri on PD4 was dissected longitudinally, rinsed three times in HBSS, and incubated in 3.5 ml of HBSS containing 100 mg/ml Trypsin and 60 mg/ml dispase at 4 °C for 1 hr, room temperature for 1 hr, and 37 °C for 10 min. The epithelium was washed out in HBSS after digested, collected, centrifuged, and precipitated. After removing the supernatant, epithelial cells were suspended in DMEM/F12 medium and adjusted to the density of 1.5×107 cells /ml. The epithelial cell suspension was mixed with ice-precooled ECM (1:3, 356231, BD biocoat, Becton-Dickinson, MA). The epithelial cells suspended in ECM were seeded onto the preheated 24-well plate and cultured in CO2 incubator at 37 °C with Organoid medium, including 1% ITS-G (PB180429, Procell, Wuhan, China), 2 mM L-glutamine (49419, Sigma-Aldrich), 1 mM nicotinamide (49419, Sigma-Aldrich), 2% B27 (17504–044, Gibco, Grand Island, NY), 1% N2 (17502–048, Gibco), 50 ng/ml EGF (HY-P7067, MedChemExpress, NJ, USA),100 ng/ml FGF-basic (HY-P7066, MedChemExpress), 100 ng/ml Noggin (HY-P70785, MedChemExpress), 200 ng/ml WNT-3A (315–20, Peprotech, Rocky Hill, USA), 200 ng/ml R-Spondin-1 (HY-P76012, MedChemExpress), and 0.5 μM A83-01 (HY-10432, MedChemExpress) in DMEM/F12. The organoids with lumen were formed after 6–7 days culture and treated with TNF (10 and 100 ng/ml) for 3 or 6 hr.
Isolation and treatment of mouse endometrial stromal cells
Request a detailed protocolMouse endometrial stromal cells were isolated as previously described (Zheng et al., 2020). Briefly, mouse uteri on PD4 were cut longitudinally, washed in HBSS, and incubated with 1% (W/V) trypsin and 6 mg/ml dispase in 3.5 mL HBSS for 1 hr at 4 °C, for 1 hr at room temperature and for 10 min at 37 °C. The uterine tissues were washed with Hanks' balanced salt solution, incubated in 6 ml of HBSS containing 0.15 mg/ml Collagenase I (Invitrogen, 17100–017) at 37 °C for 35 min. Primary endometrial stromal cells were cultured with DMEM/F12 containing 10% FBS.
Mouse endometrial stromal cells were induced for in vitro decidualization as previously described (Liang et al., 2014). Primary endometrial stromal cells were treated with 10 nM of Estradiol-17 β and 1 μM of P4 in DMEM/F12 containing 2% charcoal-treated FBS (cFBS, Biological Industries) to induce decidualization in vitro for 72 hr. Stromal cells were treated with TNC (5, 50 and 500 ng/ml, 3358-TC-050, R&D systems), S100A4 (50 and 500 ng/ml, HY-P71084, MedChemExpress), SPARC (1 and 10 μM, HY-P71086, MedChemExpress), AA (0.2, 2 and 20 μM, A3611, Sigma-Aldrich), PGI analogue ILOPROST (0.1, 1 and 10 μM, HY-A0096, MedChemExpress), SELEXIPAG (0.1, 1 and 10 μM, HY-14870, MedChemExpress), PPARδ agonist GW501516 (0.1, 1 and 10 μg/ml, 317318-70-0, Cayman Chemical, Ann Arbor, MI), COX-2 antagonist NS 398 (20 and 40 μM, S8433, Selleck, Shanghai, China), PPARδ antagonist GSK0660 (40 μM, 1014691-61-2, Selleck), and ACTIVIN A (1, 10 and 100 ng/ml, HY-P70311, MedChemExpress) in DMEM/F12 containing 2% cFBS, respectively.
Culture and treatment of human cell lines
Request a detailed protocolHuman endometrial adenocarcinoma Ishikawa cells (Chinese Academy of Science, Shanghai, China) and human endometrial stromal 4003 cells (CRL-4003, American Type Culture Collection) were cultured in DMEM/F12 with 10% FBS, and supplemented with 100 units/ml penicillin and 0.1 mg/ml streptomycin (PB180429, Procell) at 37 °C, 5% CO2, respectively. TNF (1 and 10 ng/ml) was used to treat Ishikawa cells. For further experiments, cultured 4003 cells were treated with AA (0.2, 2 and 20 μM), PGI analogue ILOPROST (0.1, 1 and 10 μM), PPARδ agonist GW501516 (0.1, 1 and 10 μg/ml) and ACTIVIN A (1, 10 and 100 ng/ml). The STR profiling of both Ishikawa cells and 4003 cells was used to confirm the identity of these cell lines. Both Ishikawa cells and 4003 cells were tested for mycoplasma-free.
Co-culture of epithelial cells and stromal cells
Request a detailed protocolHuman Ishikawa cells were cultured in 30 mm culture plates. Human 4003 cells were cultured on glass slipper. After reaching 70–80% confluence, human 4003 cells cultured on glass slippers were transferred into Ishikawa culture plates and supported with 4 prefixed plastic pillars. Human 4003 cells on glass slipper were co-cultured with the underlying Ishikawa cells in one culture plate. After the co-cultured 4003 cells and Ishikawa cells were treated with TNF, 4003 cells and Ishikawa cells were further analyzed, respectively.
Western blot
Request a detailed protocolWestern blot was performed as previously described (Li et al., 2020). The primary antibodies used in this study included CPLA2α (1:1000, SC-438, Santa Cruz Biotechnology, Dallas, TX), phosphorylated CPLA2α (1:1000), TNC (1:1000), SPARC (1:1000), α-SMA (1:1000, 19,245T, Cell Signaling Technology), COX-2 (1:1000, 12,282T, Cell Signaling Technology), PGIS (1:1000, 100023, Cayman Chemical), PPARδ (1:1000, ab178866, Abcam), BMP2 (1:1000, A0231, Abclonal, Wuhan, China), WNT4 (1:1000, sc-376279, Santa Cruz Biotechnology), E2F8 (1:1000, A1135, Abclonal), CYCLIN D3 (1:1000, 2936T, Cell Signaling Technology), TUBULIN (1:1000, 2144 S, Cell Signaling Technology), GAPDH (1:1000, sc-32233, Santa Cruz Biotechnology). After the membranes were incubated with an HRP-conjugated secondary antibody (1:5000, Invitrogen) for 1 hr, the signals were detected with an ECL Chemiluminescent Kit (Millipore, USA). Each experiment was repeated at least three times.
Real-time RT-PCR
Request a detailed protocolThe total RNA was isolated using the Trizol Reagent Kit (9109, Takara, Japan), digested with RQ1 deoxyribonuclease I (Promega, Fitchburg, WI), and reverse-transcribed into cDNA with the Prime Script Reverse Transcriptase Reagent Kit (Takara, Japan). For real-time PCR, the cDNA was amplified using a SYBR Premix Ex Taq Kit (TaKaRa) on the CFX96 Touch Real-Time System (Bio-Rad). For real-time PCR System (Bio-Rad). Data were analyzed using the 2-△△Ct method and normalized to Rpl7 (mouse) or RPL7 (human) level. The corresponding primer sequences of each gene were provided in Table 1. Each experiment was repeated at least three times.
Statistical analysis
Request a detailed protocolThe data were analyzed by GraphPad Prism8.0 Student’s T test was used to compare differences between two groups. The comparison among multiple groups was performed by ANOVA test. All the experiments were repeated independently at least three times. In the mouse study, each group had at least three mice. Data were presented as mean ± standard deviation (SD). A value of p<0.05 was considered significant.
Data availability
All data generated or analysed during this study are included in the manuscript. Source data files are provided for Figures 1-7.
References
-
Mechanosensing dysregulation in the fibroblast: A hallmark of the aging heartAgeing Research Reviews 63:101150.https://doi.org/10.1016/j.arr.2020.101150
-
Attenuation of bleomycin-induced pulmonary fibrosis by follistatinAmerican Journal of Respiratory and Critical Care Medicine 172:713–720.https://doi.org/10.1164/rccm.200412-1620OC
-
Selexipag: An oral and selective IP prostacyclin receptor agonist for the treatment of pulmonary arterial hypertensionJournal of Medicinal Chemistry 58:7128–7137.https://doi.org/10.1021/acs.jmedchem.5b00698
-
Pharmacological inhibition of autophagy by 3-MA attenuates hyperuricemic nephropathyClinical Science 132:2299–2322.https://doi.org/10.1042/CS20180563
-
Inhibition of hatching of mouse blastocysts in vitro by various prostaglandin antagonistsJournal of Reproduction and Fertility 63:359–363.https://doi.org/10.1530/jrf.0.0630359
-
Sterile inflammation: sensing and reacting to damageNature Reviews. Immunology 10:826–837.https://doi.org/10.1038/nri2873
-
Effects of androgen on embryo implantation in the mouse delayed-implantation modelFertility and Sterility 90:1376–1383.https://doi.org/10.1016/j.fertnstert.2007.07.1341
-
Cardiomyocyte ATP release through pannexin 1 aids in early fibroblast activationAmerican Journal of Physiology-Heart and Circulatory Physiology 303:H1208–H1218.https://doi.org/10.1152/ajpheart.00251.2012
-
Fibroblast activation in vascular inflammationJournal of Thrombosis and Haemostasis 9:619–626.https://doi.org/10.1111/j.1538-7836.2011.04209.x
-
Prostacyclin primes pregnant human myometrium for an enhanced contractile response in parturitionThe Journal of Clinical Investigation 118:3966–3979.https://doi.org/10.1172/JCI33800
-
Fibroblasts, myofibroblasts, and wound contractionThe Journal of Cell Biology 124:401–404.https://doi.org/10.1083/jcb.124.4.401
-
Expression of inhibin/activin subunits and their receptors and binding proteins in human preimplantation embryosJournal of Assisted Reproduction and Genetics 16:73–80.https://doi.org/10.1023/a:1022564722353
-
Intrauterine adhesion prevention after hysteroscopy: a systematic review and meta-analysisAmerican Journal of Obstetrics and Gynecology 215:267–275.https://doi.org/10.1016/j.ajog.2016.05.001
-
Evidence of prostaglandin involvement in blastocyst implantationJournal of Embryology and Experimental Morphology 55:109–122.
-
Human fallopian tubes express prostacyclin (PGI) synthase and cyclooxygenases and synthesize abundant PGIThe Journal of Clinical Endocrinology and Metabolism 87:4361–4368.https://doi.org/10.1210/jc.2002-020199
-
Proteomic analysis identifies Tenascin-C expression is upregulated in uterine fibroidsReproductive Sciences 26:476–486.https://doi.org/10.1177/1933719118773420
-
Blastocyst invasion and the stromal response in primatesHuman Reproduction 14 Suppl 2:45–55.https://doi.org/10.1093/humrep/14.suppl_2.45
-
The arachidonic acid metabolite 11,12-epoxyeicosatrienoic acid alleviates pulmonary fibrosisExperimental & Molecular Medicine 53:864–874.https://doi.org/10.1038/s12276-021-00618-7
-
Fibroblast activation in cancer: when seed fertilizes soilCell and Tissue Research 365:607–619.https://doi.org/10.1007/s00441-016-2467-x
-
Potential roles of metalloproteinases of endometrium-derived exosomes in embryo-maternal crosstalk during implantationJournal of Cellular Physiology 233:4530–4545.https://doi.org/10.1002/jcp.26259
-
Bmp2 is critical for the murine uterine decidual responseMolecular and Cellular Biology 27:5468–5478.https://doi.org/10.1128/MCB.00342-07
-
Bone morphogenetic protein 2 functions via a conserved signaling pathway involving Wnt4 to regulate uterine decidualization in the mouse and the humanThe Journal of Biological Chemistry 282:31725–31732.https://doi.org/10.1074/jbc.M704723200
-
Egr1 protein acts downstream of estrogen-leukemia inhibitory factor (LIF)-STAT3 pathway and plays a role during implantation through targeting Wnt4The Journal of Biological Chemistry 289:23534–23545.https://doi.org/10.1074/jbc.M114.588897
-
The evolution of human reproduction: a primatological perspectiveAmerican Journal of Physical Anthropology Suppl 45:59–84.https://doi.org/10.1002/ajpa.20734
-
Molecular signaling regulating endometrium-blastocyst crosstalkInternational Journal of Molecular Sciences 21:23.https://doi.org/10.3390/ijms21010023
-
cDNA cloning of a mouse prostacyclin receptor. Multiple signaling pathways and expression in thymic medullaThe Journal of Biological Chemistry 269:9986–9992.
-
TGFβ superfamily signaling and uterine decidualizationReproductive Biology and Endocrinology 15:84.https://doi.org/10.1186/s12958-017-0303-0
-
In search of definitions: Cancer-associated fibroblasts and their markersInternational Journal of Cancer 146:895–905.https://doi.org/10.1002/ijc.32193
-
The myofibroblast at a glanceJournal of Cell Science 133:jcs227900.https://doi.org/10.1242/jcs.227900
-
Follistatin attenuates early liver fibrosis: effects on hepatic stellate cell activation and hepatocyte apoptosisAmerican Journal of Physiology-Gastrointestinal and Liver Physiology 290:G137–G144.https://doi.org/10.1152/ajpgi.00080.2005
-
Fibroblast activation and senescence in oral cancerJournal of Oral Pathology & Medicine 46:82–88.https://doi.org/10.1111/jop.12456
-
Cyclical endometrial repair and regenerationDevelopment 148:dev199577.https://doi.org/10.1242/dev.199577
-
Roles of fibroblasts in microenvironment formation associated with radiation-induced cancerAdvances in Experimental Medicine and Biology 1329:239–251.https://doi.org/10.1007/978-3-030-73119-9_13
-
Cyclooxygenase isozymes: the biology of prostaglandin synthesis and inhibitionPharmacological Reviews 56:387–437.https://doi.org/10.1124/pr.56.3.3
-
Role of prostaglandins in fibroblast activation and fibrosisJournal of Cell Communication and Signaling 4:75–77.https://doi.org/10.1007/s12079-010-0089-8
-
Arachidonic acid induces differentiation of uterine stromal to decidual cellsEuropean Journal of Cell Biology 78:398–406.https://doi.org/10.1016/S0171-9335(99)80082-X
-
Myofibroblasts and mechano-regulation of connective tissue remodellingNature Reviews. Molecular Cell Biology 3:349–363.https://doi.org/10.1038/nrm809
-
Extracellular matrix and dermal fibroblast function in the healing woundAdvances in Wound Care 5:119–136.https://doi.org/10.1089/wound.2014.0561
-
Fibroblasts in the tumor microenvironmentAdvances in Experimental Medicine and Biology 1234:15–29.https://doi.org/10.1007/978-3-030-37184-5_2
-
Moesin is involved in the cytoskeletal remodelling of rat decidual cellsActa Histochemica 110:491–496.https://doi.org/10.1016/j.acthis.2008.02.002
-
Stage-specific integration of maternal and embryonic peroxisome proliferator-activated receptor δ signaling is critical to pregnancy successJournal of Biological Chemistry 282:37770–37782.https://doi.org/10.1074/jbc.M706577200
-
Hypomethylation-induced expression of S100A4 in endometrial carcinomaModern Pathology 20:1045–1054.https://doi.org/10.1038/modpathol.3800940
-
A positive feedback loop bi-stably activates fibroblastsNature Communications 9:3016.https://doi.org/10.1038/s41467-018-05274-6
-
Regulation of heterogeneous cancer-associated fibroblasts: the molecular pathology of activated signaling pathwaysJournal of Experimental & Clinical Cancer Research 39:112.https://doi.org/10.1186/s13046-020-01611-0
-
TNF-α regulated endometrial stroma secretome promotes trophoblast invasionFrontiers in Immunology 12:737401.https://doi.org/10.3389/fimmu.2021.737401
-
In situ Synthesized Monosodium Urate Crystal Enhances Endometrium Decidualization via Sterile Inflammation During PregnancyFrontiers in Cell and Developmental Biology 9:702590.https://doi.org/10.3389/fcell.2021.702590
Article and author information
Author details
Funding
National Natural Science Foundation of China (32171114 and 31871511)
- Zeng-Ming Yang
National Key Research and Development Program of China (2018YFC1004400)
- Zeng-Ming Yang
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
Funding This study was supported by National Natural Science Foundation of China (32171114 and 31871511) and National Key Research and Development Program of China (2018YFC1004400).
Ethics
All animal protocols were approved by the Animal Care and Use Committee of South China Agricultural University (No. 2021f085).
Copyright
© 2023, Chen et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 931
- views
-
- 158
- downloads
-
- 19
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 19
- citations for umbrella DOI https://doi.org/10.7554/eLife.82970






