Synchronization of oscillatory growth prepares fungal hyphae for fusion
Figures
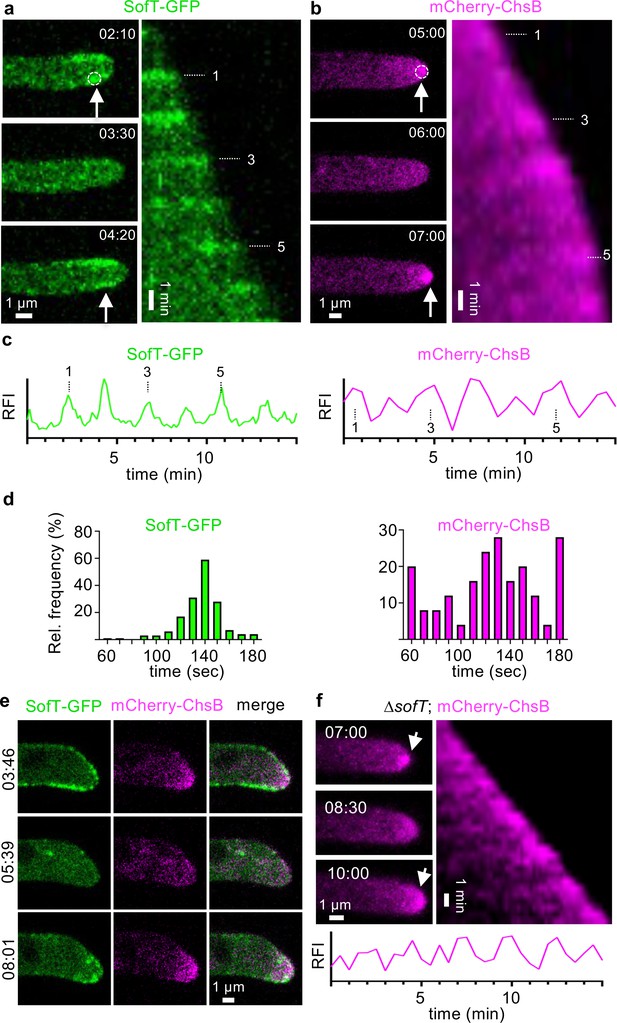
Oscillatory recruitment of signaling proteins during hyphal growth of A. flagrans.
(a, b) Time course of SofT-GFP and mCherry-ChsB localization at hyphal tips. Arrows indicate the accumulation of the proteins at the hyphal tips. Dotted circles indicate an area of 6×6 pixels which was used to measure the fluorescent intensity over time depicted in (c) of GFP-SofT at the lower edge of the plasma membrane or of mCherry-ChsB at the apex. (b) is a maximum-intensity projection of the time-lapse sequence. A kymograph was created for each time course by drawing a line (pixel width 5) along the growth axis of the respective hypha. The numbers represent the count of oscillating accumulations of each fusion protein during the growth of each hypha in the corresponding time course. (c) Relative fluorescent intensity (RFI, y-axis, arbitrary units) at the hyphal tips of a–b was measured over the time course (x-axis in minutes). (d) The interval between two peaks at hyphal tips was counted and depicted as relative frequency (y-axis) over the time (in seconds). GFP-SofT n=164 in 8 hyphae. mCherry-ChsB n=50 in 7 hyphae. (e) Localization of GFP-SofT (depicted in green) and mCherry-ChsB (depicted in magenta) during hyphal tip growth. Numbers indicate the time in minutes. (f) Localization of ChsB-mCherry at the hyphal tip of the A. flagrans ∆sofT-mutant strain. The relative fluorescent intensity at the hyphal tip (y-axis, arbitrary units) was measured over the time course.
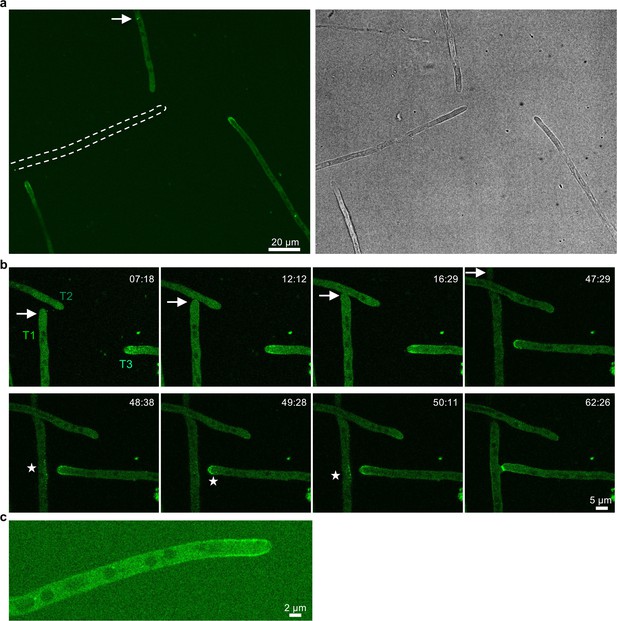
Localization of SofT-GFP at hyphal tips of A. flagrans.
(a) SofT-GFP is present in three of the four hyphae. Dashed lines outline the shape of one hypha without SofT-GFP recruitment. The arrow indicates an accumulation of SofT-GFP near the nucleus of one hypha. (b) Time course of SofT-GFP during a hyphal fusion event. The arrow indicates the hyphal tip of hypha T1. No apparent accumulation of SofT-GFP was observed during the initial phase of the time course. The star indicates accumulation of SofT-GFP in hyphae T1 and T3 at later stages of the time course. See Video 2. (c) SofT-GFP localization in a A. flagrans hypha grown on potato dextrose agar (PDA) for 24 hr.
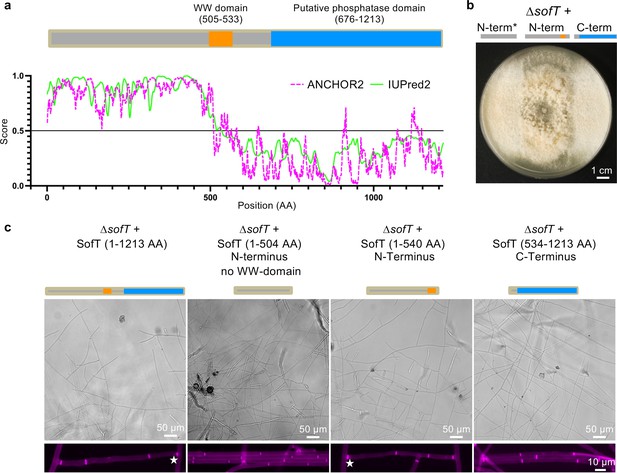
The N-terminal disordered region and WW domain of SofT are sufficient to perform vegetative hyphal fusion in A. flagrans.
(a) Domain structure of the A. flagrans SofT and prediction of disordered protein regions using iupred2a (https://iupred2a.elte.hu). The protein contains 1213 amino acids and harbors a WW domain (AA505–533) and a putative C-terminal phosphatase domain (AA676–1213). (b) Complementation experiment of the ∆sofT mutant with three truncated sofT fragments. The first fragment encoded for the N-terminal region of the protein (1–504 AA) without the conserved WW domain. The second fragment encoded for the N-terminal region with the conserved WW domain (1–540 AA). The third fragment encoded the C-terminal region of the protein containing the putative phosphatase domain (534–1213 AA). We individually transformed the three constructs into the ∆sofT strain and observed the growth of the mutants after genotyping on potato dextrose agar (PDA) after 6 days of incubation at 28°C. The ∆sofT deletion shows a strong growth phenotype on solid media (Youssar et al., 2019) with the absence of aerial mycelium. (c) Micrographs showing hyphal growth of the different rescued ∆sofT strains on low-nutrient agar (LNA) after 24 hr of incubation at 28°C. Stars indicate hyphal fusion events. A rescue with the full-length sofT construct was used as a control.
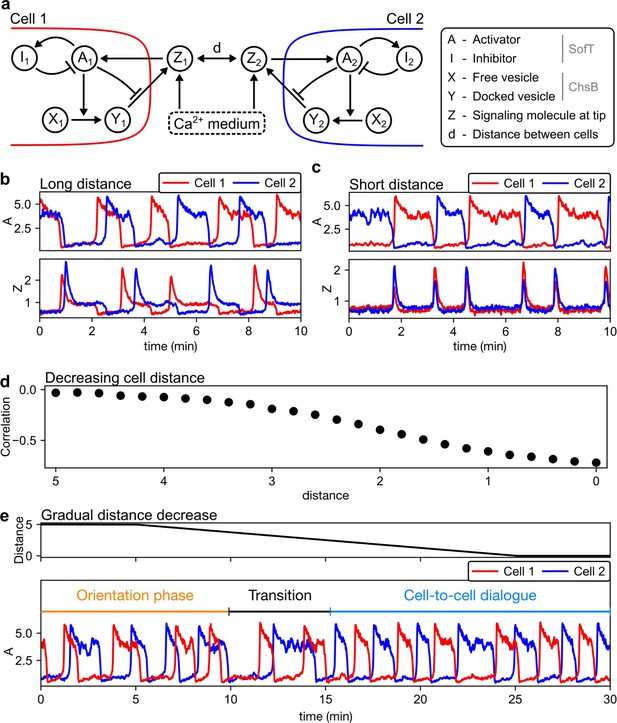
A mathematical model demonstrates the emergence of the temporal synchronization of two hyphae approaching each other.
(a) In the model, each hyphal cell contains an excitatory system with an activator () and inhibitor (). This excitatory system is triggered by extracellular signaling molecules () and, in turn, regulates the docking and subsequent release of vesicles ( and ), which contain the signaling protein, into the extracellular space. (b) At long distances (here, ), the cells operate as independent oscillator units. (c) At short distances (here, ), cells synchronize and oscillate in anti-phase. (d) Decreasing the distance between the two cells increases the magnitude of the anti-correlation (negative Pearson correlation coefficient) between the activator concentrations of the individual cells. (e) Upon continuous reduction of the distance between the cells, anti-synchrony emerges (noticeable at around 15 min).
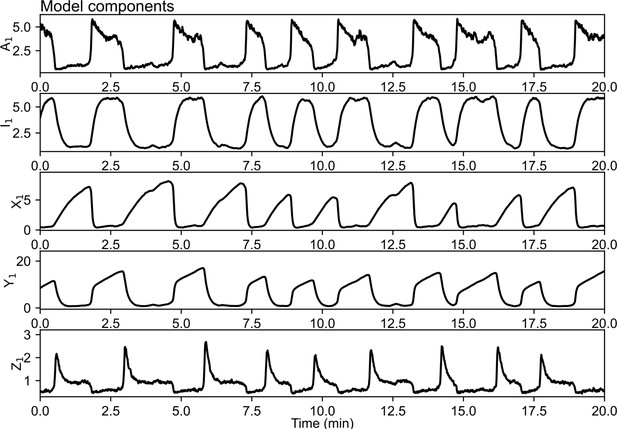
Example simulation containing a single cell to illustrate the oscillatory dynamics of all model variables.
To illustrate the time course of all model variables, a simulation with a single cell was implemented by setting the cell distance to infinity ().
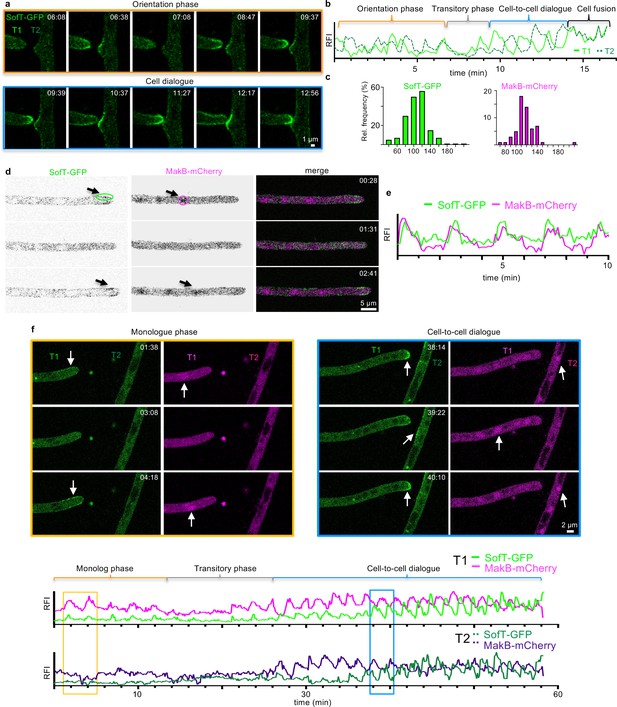
Decoupling of of SofT and MakB oscillations entering the cell-to-cell dialogue.
(a, b) Time course of SofT-GFP during a hyphal fusion event. Selective micrographs of the time course are shown and are divided into an orientation phase (orange frame) and a cell-to-cell dialogue phase (blue frame). An area of 3×3 pixels at each hyphal apex was used to measure the fluorescent intensity depicted in (b) (y-axis, arbitrary units). T1=left tip; T2=right tip. (c) The interval between two peaks of SofT-GFP or MakB-mCherry at each hyphal tip during the cell dialogue was counted and the distribution is depicted as relative frequency (y-axis) over the time (in seconds). SofT-GFP n=178 in 18 hyphae. MakB-mCherry n=57 in 8 hyphae. (d) Co-localization of GFP-SofT (depicted in green) and MakB-mCherry (depicted in magenta) during hyphal tip growth. Arrows indicate the accumulation of the proteins at the hyphal tips. The circles indicate an area of 15×4 (SofT) and 10×10 (MakB) pixels which were used to measure the fluorescent intensity over time depicted in (e) of GFP-SofT at the upper edge of the plasma membrane or of MakB-mCherry in the nucleus. Single channels are depicted as inverted grayscale. (e) Relative fluorescent intensity (RFI, y-axis, arbitrary units) at the hyphal tips of (d) was measured over the time course (x-axis in minutes). (f) Time course of SofT-GFP and MakB-mCherry during a hyphal fusion event. Selective micrographs of the time course are shown and are divided into a monologue phase (orange frame) and cell-to-cell dialogue phase (blue frame). T1=left tip; T2=right hypha. Arrows indicate the accumulation of the proteins in the hyphae. An area of 21×21 pixels was used to measure the fluorescent intensity of MakB-mCherry, an area of 10×6 pixels was used to measure the fluorescent intensity of SofT-GFP depicted in the lower graphs (y-axis, arbitrary units). Each graph shows the fluorescence of one hypha.
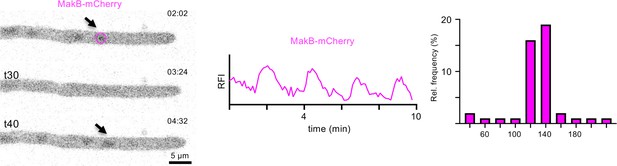
Oscillatory recruitment of MakB-mCherry during hyphal growth of A. flagrans.
Time course of MakB-mCherry at a hyphal tip. Arrows indicate the accumulation of the protein at the nucleus. The circle indicates an area of 15×15 pixels which was used to measure the fluorescent intensity over time. The representative images are a maximum-intensity projection of the time-lapse sequence. The relative fluorescent intensity (RFI, y-axis, arbitrary units) at the region of interest was measured over the time course (x-axis in minutes) and is depicted as graph. The interval between two peaks at the region of interest was counted and depicted as relative frequency (y-axis) over the time (in seconds). n=45 in 8 hyphae. See Video 6.
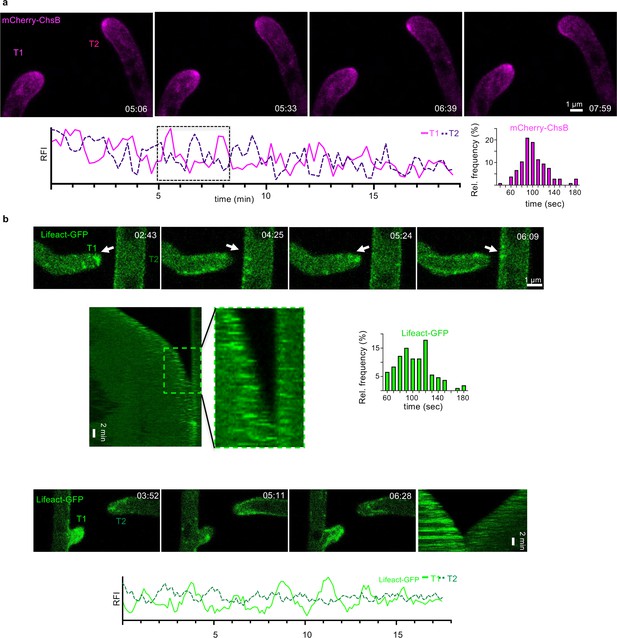
The oscillating extension of hyphae is synchronized during hyphal fusion in A.flagrans.
(a) Time course of mCherry-ChsB during a hyphal fusion event. A maximum-intensity projection was generated from the time-lapse sequence. It was further bleach-corrected using the bleach correction plugin (correction method: simple ratio) of Fiji. The relative fluorescent intensity (RFI) of an area (y-axis, arbitrary units) of 6×6 pixel was measured at each hyphal apex over time (x-axis in minutes) to generate the graph. The boxed area depicts the selected micrographs of the time course. The interval between two peaks of ChsB at each hyphal tip during the cell-to-cell dialogue was counted and the distribution is depicted as relative frequency (y-axis) over the time (in seconds). n=105 in 11 hyphae. (b) Maximum-intensity projections of two time courses of Lifeact-GFP during hyphal fusion events. A maximum-intensity projection was generated from each time-lapse sequence. Arrows indicate the accumulation of the protein in the hyphae. Kymographs were created of the time courses by drawing a line (pixel width 5) along the growth axis of the respective hyphae. The boxed area of the kymograph is enlarged and displays the phase of the cell-to-cell dialogue. The interval between two peaks of Lifeact-GFP at each hyphal tip during the cell-to-cell dialogue was counted and the distribution is depicted as relative frequency (y-axis) over the time (in seconds). n=106 in 10 hyphae. See Video 9.
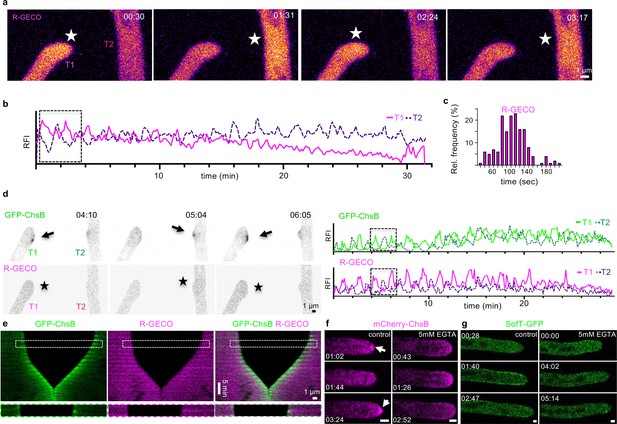
The stepwise extension of hyphae is coordinated during the cell-to-cell dialogue stage of hyphal fusion.
Maximum-intensity projection of a time course of R-GECO during a hyphal fusion event. Stars indicate the fluorescent excitation of R-GECO in the presence of Ca2+ inside the hyphae. The changes in fluorescent intensity are color-coded as Fire LUT using Fiji, depicting high pixel values as white and yellow color, and low pixel values as blue and magenta. The relative fluorescent intensity (RFI) (y-axis, arbitrary units) was measured at each hyphal tip over the time course (x-axis in minutes). The RFI of an area of 12×12 pixels was measured at each hyphal tip to generate the graph. The boxed area depicts the selected micrographs of the time course. (c) The interval between two peaks of R-GECO at each hyphal tip during the cell-to-cell dialogue was counted and the distribution is depicted as relative frequency (y-axis) over the time (in seconds). n=160 in 24 hyphae. (d) Maximum-intensity projection of a time course of GFP-ChsB and R-GECO during a hyphal fusion event. Single channels are depicted as inverted grayscale. Arrows indicate the localization of GFP-ChsB at hyphal tips. Stars indicate the fluorescent excitation of R-GECO in the presence of Ca2+ inside the hyphae. The RFI (y-axis) was measured of an area of 6×6 pixels at each hyphal apex over the time course (x-axis in minutes, y-axis, arbitrary units). The boxed area of the selected micrographs represents the enlarged area in (e). (e) A kymograph was created of the time course in (d) drawing a line (pixel width 5) along the growth axis of both hyphae. The boxed area is enlarged and displays one cycle of the cell-to-cell dialogue. (f) Compared to the control (low-nutrient agar [LNA] containing 1 µM CaCl2), mCherry-ChsB did not accumulate at the Spitzenkörper on LNA containing 5 mM EGTA. (g) Compared to the control, GFP-SofT did not accumulate at hyphal tips on LNA containing 5 mM EGTA. Scale bars in (f, g) depict 1 µm.
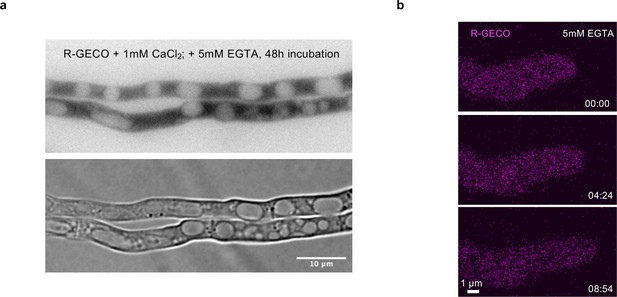
Extracellular Ca2+ is necessary for hyphal fusion and pulse-like exocytosis.
(a) Hyphae showed close physical contact without hyphal fusion after incubation with the Ca2+ chelating agent EGTA (5 mM). (b) No pulses of R-GECO were observed in hyphae on low-nutrient agar (LNA) containing 5 mM EGTA.
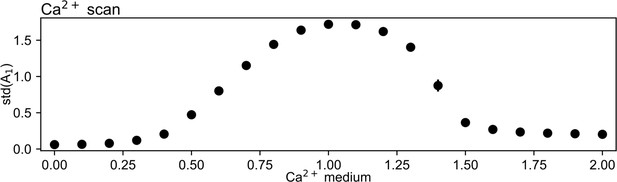
Sufficient concentration of Ca2+ is necessary to produce oscillations.
In a simulation containing only a single cell, at a Ca2+ concentration of 0 (), no oscillations are observed. Around a Ca2+ concentration of 1 (), the cell generates regular oscillations (as shown in Figure 1), evidenced by an increasing standard deviation of the activator level (). Increasing the Ca2+ concentration further leads again to the gradual loss of oscillatory behavior. The standard deviation was calculated over a simulation that represented 800 min of cellular dynamics.
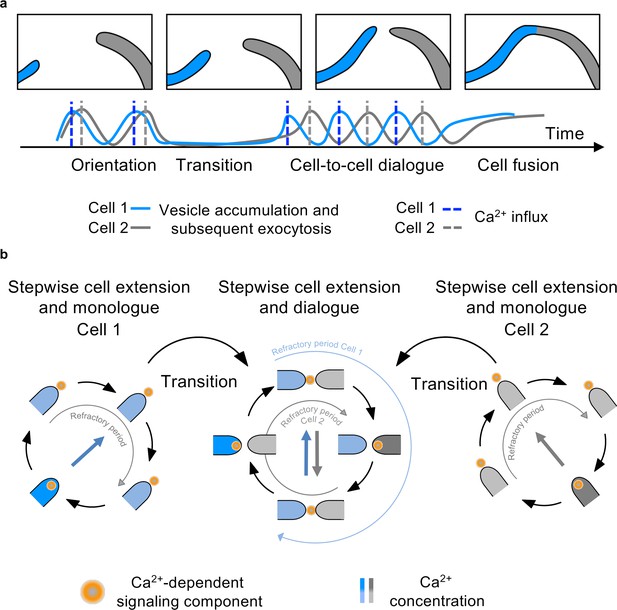
Scheme of the cell-to-cell dialogue.
(a) Cell fusion of two hyphal cells is mediated by the synchronization of oscillatory growth. Vesicles needed for hyphal growth and communication accumulate at the hyphal tip and are released to the surroundings upon an influx of Ca2+. If two cells are in close proximity, the uncoordinated, stepwise growth shifts after transitory entrainment to a synchronized anti-phasic cell dialogue and subsequent cell fusion. (b) During stepwise cell extension, a Ca2+-dependent signaling component is released with a refractory period to prevent self-stimulation. Entrainment of two cells in close proximity initiates a cell dialogue mediated by a so far unknown Ca2+-dependent signaling component. Refractory periods after secretion prevent self-stimulation.
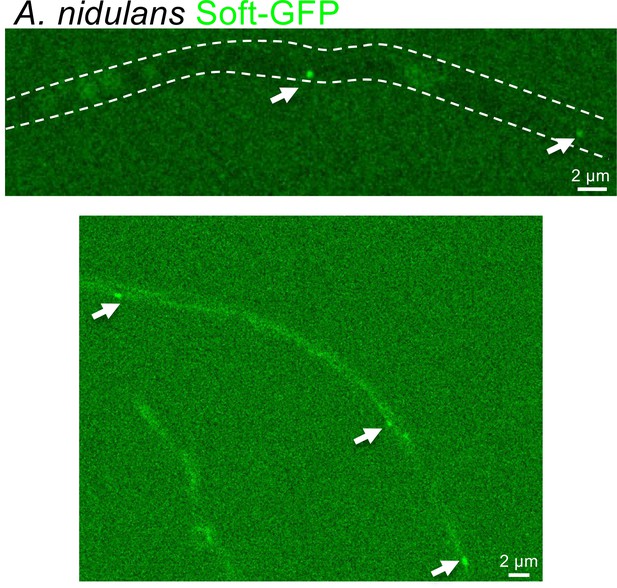
Localization of Soft-GFP in A. nidulans.
Arrows indicate accumulations of Soft-GFP inside hyphae. (Top) Dashed lines outline the hypha grown on low-nutrient agar (LNA) for 16 hr at 37°C. (Bottom) Hyphae were grown on Aspergillus minimal medium for 16 hr at 37°C.
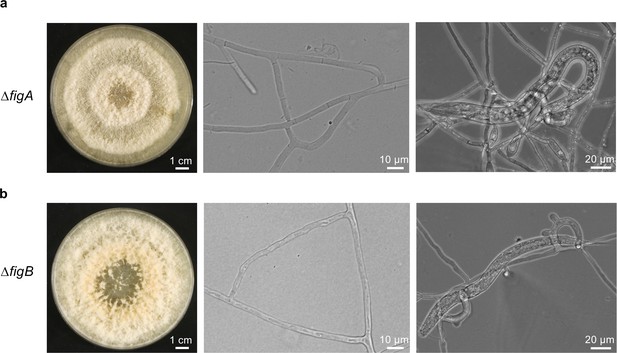
Gene deletion of figA and figB in A. flagrans.
(a) The A. flagrans ∆figA gene deletion mutant can perform vegetative hyphal fusion and trap C. elegans. (b) The A. flagrans ∆figB gene deletion mutant can perform vegetative hyphal fusion and trap C. elegans. The growth of both mutants on potato dextrose agar (PDA) is shown after 7 days at 28°C.
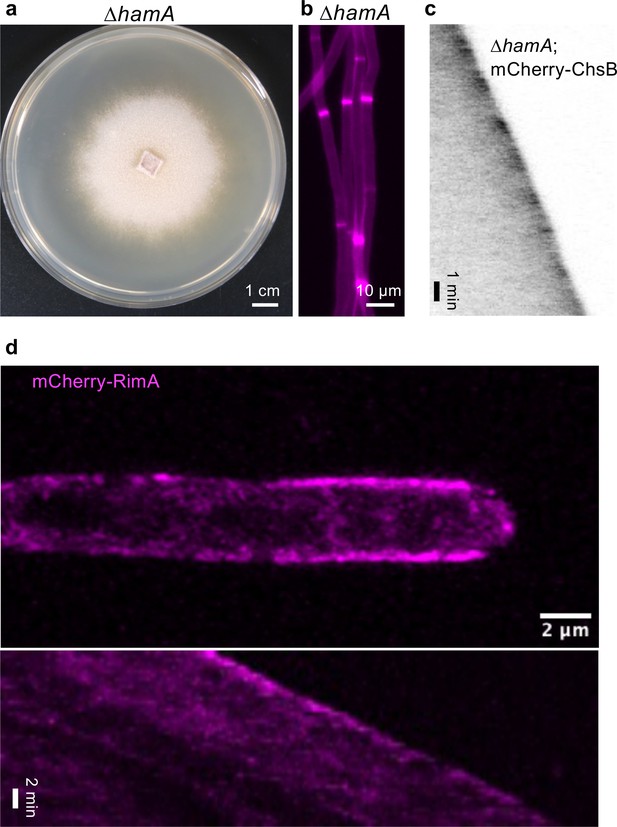
Gene deletion of hamA leads to loss of hyphal fusion.
(a) Growth of the A. flagrans ∆hamA gene deletion mutant grown on potato dextrose agar (PDA) after 7 days at 28°C. (b) Deletion of hamA leads to loss of vegetative hyphal fusion. (c) Kymograph of a time course depicts the recruitment of mCherry-ChsB the hyphal tip of a ∆hamA hypha. (d) Localization of mCherry-RimA at the hyphal tip. A maximum-intensity projection was generated from the time-lapse sequence. It was further bleach-corrected using the bleach correction plugin (correction method: simple ratio) of Fiji. A kymograph was created by drawing a line (pixel width 5) along the growth axis of the hypha. See Video 12.
Videos
Localization of GFP-SofT during hyphal tip growth.
Time course of GFP-SofT during a hyphal fusion event.
Localization of mCherry-ChsB during hyphal tip growth.
Co-localization of GFP-SofT (depicted in green) and mCherry-ChsB (depicted in magenta) during hyphal tip growth.
Time course of GFP-SofT during a hyphal fusion event.
Time course of MakB-mCherry during hyphal tip growth.
Co-localization of GFP-SofT (depicted in green) and MakB-mCherry (depicted in magenta) during hyphal tip growth.
Co-localization of GFP-SofT (depicted in green) and MakB-mCherry (depicted in magenta) during a hyphal fusion event.
Time course of Lifeact-GFP during a hyphal fusion event.
Time course of R-GECO during a hyphal fusion event.
Time course of GFP-ChsB (depicted in green) and R-GECO (depicted in magenta) during a hyphal fusion event.
Time course of mCherry-RimA during hyphal tip growth.
Tables
A. flagrans and A. nidulans strains used in this study.
| Strain number | Genotype | Origin |
|---|---|---|
| CBS 349.94 | Wild type | Youssar et al., 2019 |
| sVW16 | tubA(p)::lifeact::GFP::gluC(t); trpC(p)::hph::trpC(t) | Wernet et al., 2022 |
| sVW01 | sofT::hph (∆soft) | Youssar et al., 2019 |
| sVW20 | ∆sofT; sofT(p)::sofT::gluC(t); neo; makB(p)::makB::makB(t); ntc | Haj Hammadeh et al., 2022 |
| sVW25 | tubA(p)::lifeact::GFP::gluC(t); trpC(p)::hph::trpC(t) | Wernet et al., 2022 |
| sVW29 | sofT(p)::sofT::GFP::sofT(t); trpC(p)::hph::trpC(t) | This study |
| sVW40 | soft(p)::sofT (∆1–504 AA N-terminus) gpdA(p)::neo::trpC(t) | This study |
| sVW41 | soft(p)::sofT (∆534–1213 AA C-terminus) gpdA(p)::neo::trpC(t) | This study |
| sVW43 | soft(p)::sofT (∆1–540 AA N-terminus, no WW domain) gpdA(p)::neo::trpC(t) | This study |
| sVW51 | tubA(p)::mCherry::chsB::chsB(t); gpdA(p)::neo::trpC(t) | This study |
| sVW52 | sofT(p)::GFP::sofT; tubA(p)::mCherry::chsB::chsB(t);trpC(p)::hph::trpC(t); gpdA(p)::neo::trpC(t) | This study |
| sVW65 | h2b(p)::r-GECO::gluC(t); gpdA(p)::neo::trpC(t) | This study |
| sVW69 | h2b(p)::r-GECO::gluC(t);tubA(p)::GFP::chsB::chsB(t); gpdA(p)::neo::trpC(t); trpC(p)::hph::trpC(t) | This study |
| sVW75 | sofT::hph (∆soft); tubA(p)::mCherry::chsB::chsB(t); gpdA(p)::neo::trpC(t); trpC(p)::hph::trpC(t) | This study |
| sVW76 | figA::hph (∆figA) | This study |
| sVW77 | figB::hph (∆figB) | This study |
| sVW78 | tubA(p)::mCherry::rimA::chsB(t); gpdA(p)::neo::trpC(t) | This study |
| sVW44 | hamA::hph (∆hamA), trpC(p)::hph::trpC(t) | This study |
| sVW80 | sVW51, hamA::hph (∆hamA), trpC(p)::hph::trpC(t) | This study |
| RPA 33 | yA1, riboB2, pyrG89, pyroA4, pabaA1, nkuA::argB+ | |
| VW-a01 | yA1, riboB2, pyrG89, (AnSoft-GFP::Afpyro) pyroA4, pabaA1, nkuA::argB+ | This study |
Plasmids used in this study.
| Name | Description/genotype | Reference |
|---|---|---|
| pVW57 | Plasmid backbone containing trpC(p)::hph::trpC(t) | Wernet et al., 2022 |
| pVW92 | Plasmid backbone containing gpdA(p)::neo::trpC(t) and tubA(p) | Wernet et al., 2021 |
| pNH57 | Plasmid containing GFP::hph | Nicole Wernet (Karlsruhe) |
| pVW106 | sofT(p)::sofT::GFP::sofT(t); trpC(p)::hph::trpC(t) | This study |
| pVW118 | tubA(p)::mCherry::chsB::chsB(t);gpdA(p)::neo::trpC(t) | This study |
| pVW132 | tubA(p)::GFP::chsB::chsB(t); gpdA(p)::neo::trpC(t) | This study |
| pVW120 | tubA(p)::r-GECO::gluC(t); gpdA(p)::neo::trpC(t) | This study |
| pVW125 | h2b(p)::r-GECO::gluC(t); trpC(p)::hph::trpC(t) | This study |
| pVW131 | h2b(p)::r-GECO::trpC(t); trpC(p)::hph::trpC(t) | This study |
| pVW132 | figA::hph (∆figA) | This study |
| pVW133 | figB::hph (∆figB) | This study |
| pVW134 | tubA(p)::mCherry::rimA::chsB(t); gpdA(p)::neo::trpC(t) | This study |
| pVW113 | hamA::hph (∆hamA) | This study |
| pVW-a68 | AnSoft-GFP::Afpyro | This study |
| pVW108 | soft(p)::sofT (∆1–504 AA N-terminus) gpdA(p)::neo::trpC(t) | This study |
| pVW109 | soft(p)::sofT (∆534–1213 AA C-terminus) gpdA(p)::neo::trpC(t) | This study |
| pVW112 | soft(p)::sofT (∆1–540 AA N-terminus, no WW domain) gpdA(p)::neo::trpC(t) | This study |
Oligonucleotides used in this study.
| Name | Sequence (5’ to 3’) | Description |
|---|---|---|
| chsB_fwd | gaatggatgaactctacaaa atggcacagcaaggaggtt | mCherry-ChsB |
| chsB_rev | aggagatcttctagaaagatgatggggcgttaaggtttc | mCherry-ChsB |
| pJet_fwd | atctttctagaagatctcctacaatattc | mCherry-ChsB |
| mCherry_rev | tttgtagagttcatccattccac | mCherry-ChsB |
| tubP_rev | gatgaattatatttcgtcaagaag | GFP-ChsB; tubA(p)::r-geco |
| chsb_fwd | atggcacagcaaggaggtt | GFP-ChsB |
| gfp_chsb_fwd | ttgacgaaatataattcatcatggtttccaagggtgagg | GFP-ChsB |
| gfp_chsb_rev | taacctccttgctgtgccatagcggccgctttgtaaagtt | GFP-ChsB |
| R_geco_pOL_fwd | ttgacgaaatataattcatcatggtcgactcatcacgtc | R-GECO (tubA(p)) |
| r-geco_tOL_rev | atacatcttatctacatacgctacttcgctgtcatcatttg | R-GECO (tubA(p)) |
| tgluC_gOL_fwd | aaatgatgacagcgaagtagcgtatgtagataagatgtatgattag | R-GECO (tubA(p)) |
| tgluC_geco_rev | aggagatcttctagaaagatatcttgttggggggaaggg | R-GECO (tubA(p)) |
| h2b_p_trpCOL_fwd | ctttccctaaactccccccaggagaagaaaggagcaaaatc | R-GECO (h2b(p)) |
| h2b_p_gecoOL_rev | cgacgtgatgagtcgaccattttgaaatttgttttttgtttgggtag | R-GECO (h2b(p)) |
| trpC_rev | tggggggagtttagggaaag | R-GECO (h2b(p)) |
| r_geco_fwd | atggtcgactcatcacgtc | R-GECO (h2b(p)) |
| soft_gfp_locus_fwd | ctcgagtttttcagcaagattaccgtcctcagtacaacatg | SofT-GFP |
| soft_gfp_locus_rev | acctcacccttggaaaccatatacccatactcgcatctgg | SofT-GFP |
| soft_GFPcas_fwd | ccagatgcgagtatgggtatatggtttccaagggtgagg | SofT-GFP |
| soft_GFPcas_rev | caaccgcccggacgaatcattggggggagtttagggaaag | SofT-GFP |
| soft_Term_fwd | ctttccctaaactccccccaatgattcgtccgggcggtt | SofT-GFP |
| softterm_gfp_rev | attgtaggagatcttctagaaagattgggacgagtgggatttaaaatgga | SofT-GFP |
| figA_lb_fwd | ctcgagtttttcagcaagatTGTCGCTTGGGCTTGATAG | Deletion figA |
| figA_lb_rev | CCTCCACTAGCATTACACTTATCTGCTAACGTAACTAGACG | Deletion figA |
| figA_hph_fwd | GTCTAGTTACGTTAGCAGATAAGTGTAATGCTAGTGGAGG | Deletion figA |
| figA_hph_rev | CTAACAGGCCTATCGGAGTTTGGGGGGAGTTTAGGGAAAG | Deletion figA |
| figA_rb_fwd | CTTTCCCTAAACTCCCCCCAAACTCCGATAGGCCTGTTAG | Deletion figA |
| figA_rb_rev | aggagatcttctagaaagatCGGAGGTCGTCAAGAAGC | Deletion figA |
| figA_up_fwd | AGGAAGACCGATTACGAAAC | Deletion figA |
| figA_down_rev | CGATATACGATCCGAAGGTC | Deletion figA |
| figA_lb_g418_rev | AATGCAATGTAATAGATACCATCTGCTAACGTAACTAGACG | Deletion figA |
| figA_g418_fwd | GTCTAGTTACGTTAGCAGATGGTATCTATTACATTGCATTGCG | Deletion figA |
| figB_lb_fwd | ctcgagtttttcagcaagatGGCGAAGAGACTGGATTTATC | Deletion figB |
| figB_lb_rev | CCTCCACTAGCATTACACTTTTTGAAGTTTTGCTGATGATGTGAG | Deletion figB |
| figB_hph_fwd | ATCATCAGCAAAACTTCAAAAAGTGTAATGCTAGTGGAGGT | Deletion figB |
| figB_hph_rev | TTGTTCAGCTTTTTTCCCATTGGGGGGAGTTTAGGGAAAG | Deletion figB |
| figB_rb_fwd | CTTTCCCTAAACTCCCCCCAATGGGAAAAAAGCTGAACAAAAAAAATC | Deletion figB |
| figB_rb_rev | aggagatcttctagaaagatCGCCTGTGTAACGGCTTTTG | Deletion figB |
| figB_up_fwd | AGAGCCGCATGGTTTATTTAG | Deletion figB |
| figB_down_rev | AGCACAGAGTAACCTGGAC | Deletion figB |
| rimA_orf_fwd | GAATGGATGAACTCTACAAAATGGAAACCCCAGCTCCAG | mCherry-RimA |
| rimA_rb_rev | AGGAGATCTTCTAGAAAGATCGTTCTATGCCTGAAATCGG | mCherry-RimA |
| ham10_lb_fwd | ctcgagtttttcagcaagatGGCGGATATCAATCTTATCTTG | Deletion hamA |
| ham10_lb_rev | CCTCCACTAGCATTACACTTGGTGACCGAAATCGCCTTAT | Deletion hamA |
| ham10_hph_fwd | ATAAGGCGATTTCGGTCACCAAGTGTAATGCTAGTGGAGG | Deletion hamA |
| ham10_hph_rev | CACAAGGATGGCTTCCCATTTGGGGGGAGTTTAGGGAAAG | Deletion hamA |
| ham10_rb_fwd | CTTTCCCTAAACTCCCCCCAAATGGGAAGCCATCCTTGTG | Deletion hamA |
| ham10_rb_rev | aggagatcttctagaaagatCGTTCCGATTTACTCGTCG | Deletion hamA |
| ham10_up_fwd | GCCGAGACTACTAGCTAGG | Deletion hamA |
| ham10_down_rev | CTATATCGTTGGTTCGAGGG | Deletion hamA |
| soft_Nterm_tgluCOL_rev | ATACATCTTATCTACATACGTTAAATACCGGTCTCCGGTGC | N-terminal SofT truncation |
| soft_ntermnoWW_tgluCol_rev | ATACATCTTATCTACATACGTTATGGAAGAGGTGGGGGAGA | N-terminal SofT truncation, no WW domain |
| softProm_trpCOL_fwd | CTTTCCCTAAACTCCCCCCACAGAGTTCGAATAGCGTTGC | C-terminal SofT truncation |
| softProm_CtermOL_rev | ATACCGGTCTCCGGTGCCATTGTGGAGACGAAGGCAAAG | C-terminal SofT truncation |
| softCterm_fwd | CCTTTGCCTTCGTCTCCACAATGGCACCGGAGACCGGTATT | C-terminal SofT truncation |
| softCterm_tgluC_rev | ATACATCTTATCTACATACGTTAATACCCATACTCGCATCTG | C-terminal SofT truncation |
| VW230 | TTGTAAAACGACGGCCAGTGACACCAGAAATCTCTCCGAATG | Soft-GFP, A. nidulans |
| VW231 | GTGAAAAGTTCTTCTCCTTTCTTCCCATGCTCTAAACTCG | Soft-GFP, A. nidulans |
| VW232 | CGAGTTTAGAGCATGGGAAGAAAGGAGAAGAACTTTTCACTGG | Soft-GFP, A. nidulans |
| VW233 | TTAAAAAGACTCGGCATCAAgcgagtgtctacataatgaagg | Soft-GFP, A. nidulans |
| VW234 | ttcattatgtagacactcgcTTGATGCCGAGTCTTTTTAATGT | Soft-GFP, A. nidulans |
| VW235 | GACCATGATTACGCCAagctGGGTGACGGATTATTACCTCT | Soft-GFP, A. nidulans |





