Inhibition of mitochondrial protein import and proteostasis by a pro-apoptotic lipid
Figures
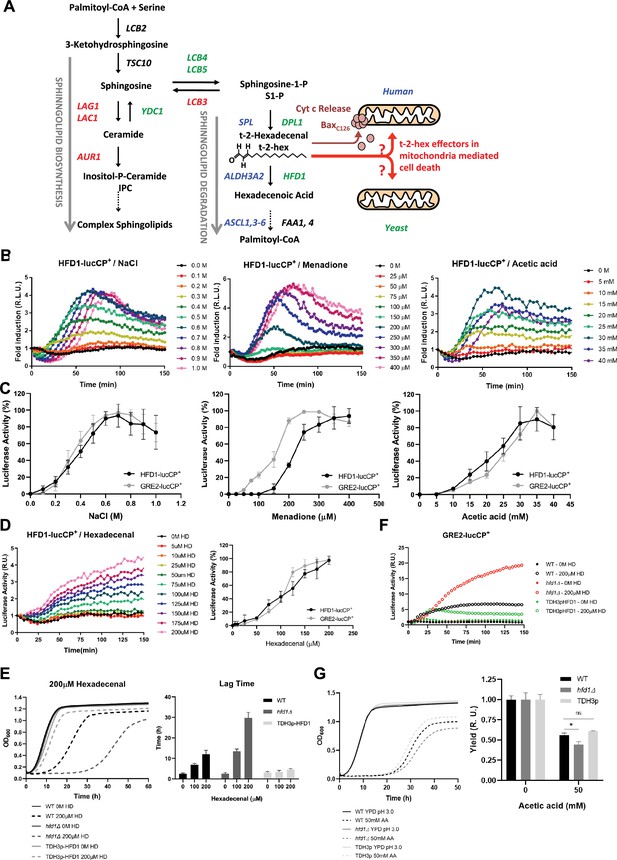
The stress-activated fatty aldehyde dehydrogenase Hfd1 determines trans-2-hexadecenal (t-2-hex) mediated toxicity and adaptive response.
(A) Overview of the generation of t-2-hex within the sphingolipid degradation pathway. Stress-activated (-repressed) enzymatic functions are depicted in green (red) for yeast. The corresponding human enzymes involved in t-2-hex chemistry are shown in blue. t-2-hex stimulates the pro-apoptotic activity of Bax at human mitochondria by C126 lipidation. (B) HFD1 expression is activated by different cytotoxic stresses. Hyperosmotic (NaCl), oxidative (Menadione), and pro-apoptotic (Acetic acid) stresses were gradually applied to HFD1p-luciferase reporter containing yeast cells. The relative luciferase activity was measured in vivo (n=3). (C) Comparison of different stress sensitivities of the HFD1 and GRE2 promoters. (D) External addition of t-2-hex (HD) activates HFD1 expression in a concentration-dependent manner. Upper panel: HFD1-luciferase activation upon gradual addition of t-2-hex (n=3). Lower panel: Comparison of HFD1 and GRE2 activation sensitivities to t-2-hex. (E) Left panel: HFD1 gene dose modulates susceptibility to t-2-hex. Growth curves of wild-type, hfd1Δ, and constitutively Hfd1 overexpressing strains (TDH3p-HFD1) in the absence and presence of t-2-hex (n=3). Right panel: Quantitative comparison of growth performance (length of lag phase) of the same strains upon the indicated t-2-hex concentrations (n=3). (F) Loss of Hfd1 function causes a hypersensitive t-2-hex response. GRE2p-luciferase reporter assay in strains with the indicated HFD1 gene dose in response to t-2-hex (n=3). (G) HFD1 gene dose modulates the tolerance to pro-apoptotic concentrations of acetic acid (n=3). *p<0.05 by Student´s unpaired t-test.
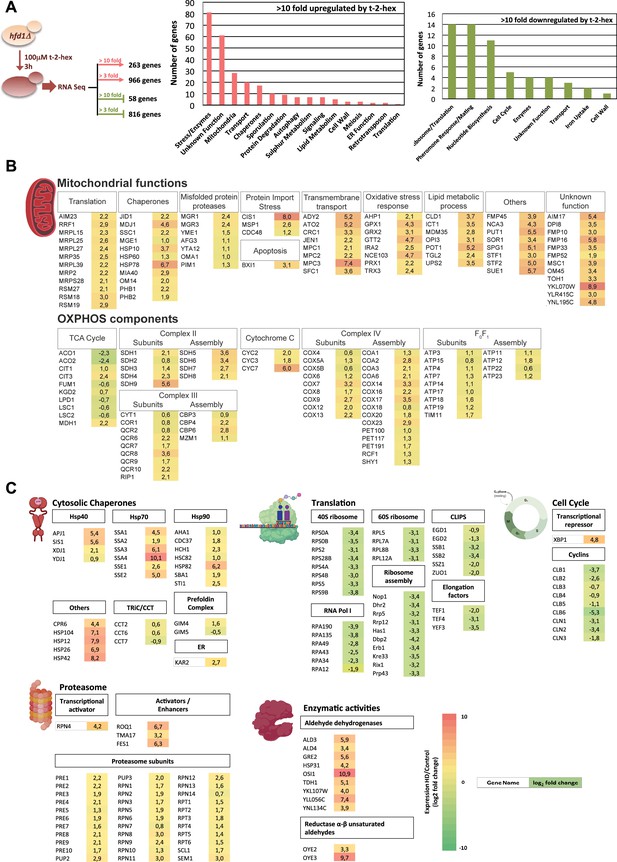
Trans-2-hexadecenal (t-2-hex) stress causes profound transcriptomic remodeling.
(A) Transcriptomic analysis of the cellular response to t-2-hex overload. RNA seq experiment setup and general functional groups of up- and down-regulated genes. (B) t-2-hex activated mitochondrial functions. (C) Genomic remodeling of gene expression reveals a general proteostatic response of the cell to t-2-hex. Cytosolic chaperones are strongly activated in general, but not the Cct chaperonin or the prefoldin complex. Structural components and regulators of the proteasome are coordinately up-regulated, while the cytosolic translation machinery and cell cycle regulators are strongly repressed. Significantly up-regulated enzymes with known aldehyde dehydrogenase or unsaturated aldehyde reductase activities are shown. Colors indicate log2 fold changes in gene expression of t-2-hex treated versus mock-treated cells (means of n=3 independent biological replicates) for genes representing selected functional groups or complexes.
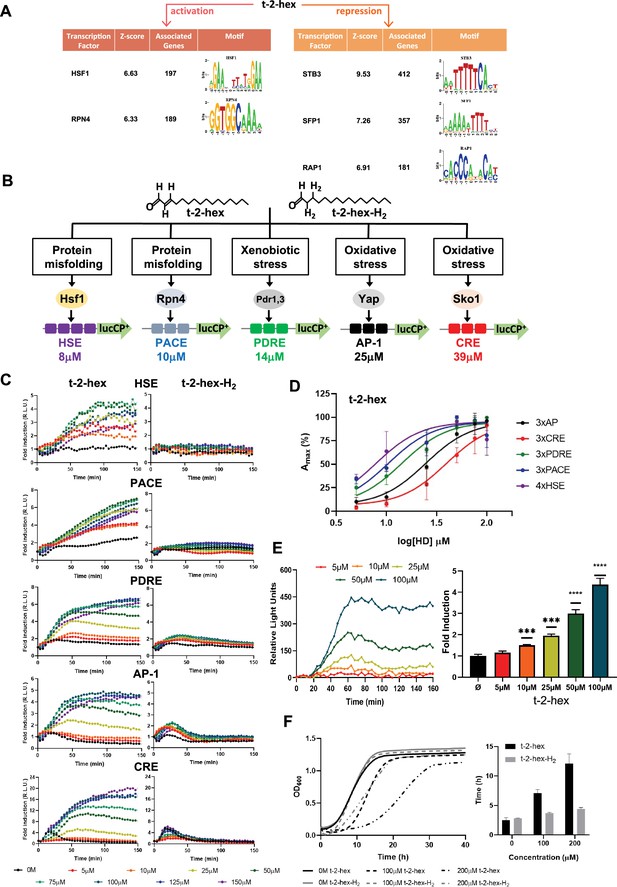
Trans-2-hexadecenal (t-2-hex) specifically induces the heat shock and proteasomal transcriptional response.
(A) Significantly enriched promoter motifs were identified from the transcriptionally up- and down-regulated genes identified by our RNA seq study upon t-2-hex stress. (B) Schematic representation of the stress-specific luciferase reporters applied in the dose-response experiments upon unsaturated t-2-hex or saturated analog t-2-hex-H2. The t-2-hex dose causing half-maximal induction for each reporter is given in μM below the constructs. (C) Dose-response curves of the indicated live cell luciferase reporters upon increasing t-2-hex and t-2-hex-H2 concentrations (n=3). All reporters were assayed in hfd1Δ cells. Initial light emission levels at time 0 were set to 1. (D) Comparison of the sensitivities of different stress type-specific responses to the pro-apoptotic t-2-hex. Experimental data from (C) were analyzed by plotting the maximal reporter activation against the log[t-2-hex] concentration. (E) Induction of a 3xPACE-lucCP+ reporter in wild-type yeast cells upon the indicated t-2-hex doses. Left panel: Dose-response curves corrected for the mock-treated samples. Right panel: Maximal induction fold for each lipid dose tested (n=3). ***p<0.001, ****p<0.0005 by Student’s unpaired t-test. (F) t-2-hex unsaturation is the cause of its severe growth inhibition. Growth of yeast wild-type cells was scored upon the indicated t-2-hex and t-2-hex-H2 concentrations (upper panel) and the lag time was calculated (lower panel), n=3.
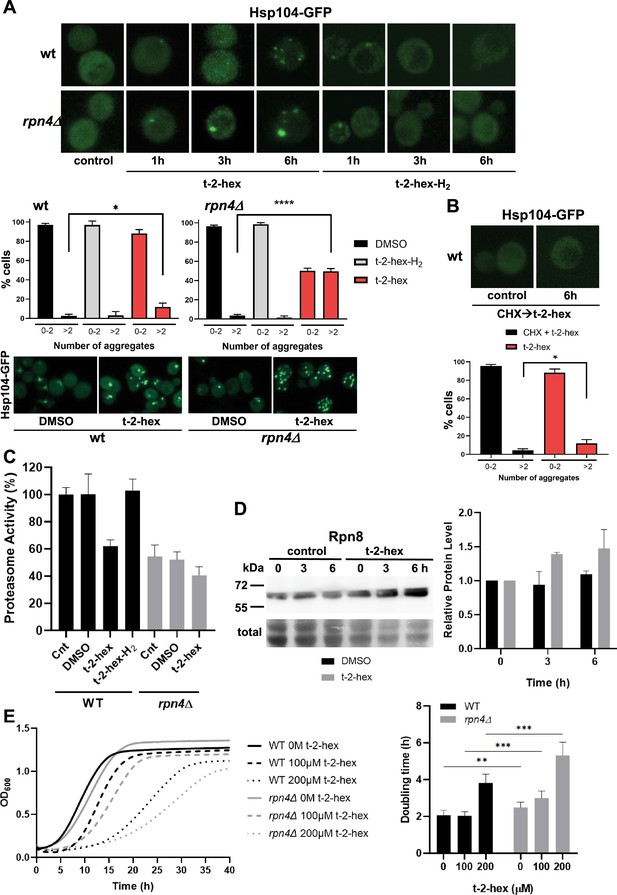
Trans-2-hexadecenal (t-2-hex) leads to cytosolic protein aggregation and inhibition of proteasomal function.
(A) GFP-tagged Hsp104 was used to visualize intracellular protein aggregation in wild-type and proteasome-deficient rpn4Δ cells. Specifically, unsaturated t-2-hex caused a slowly increasing protein aggregation. Cells were treated with 100 μM of the bioactive lipids for the indicated times. Lower panels: Quantification of t-2-hex-induced protein aggregates across cell populations. Number of analyzed cells: wt DMSO n=601, wt t-2-hex n=584, wt t-2-hex-H2 n=558; rpn4 DMSO n=551, rpn4 t-2-hex n=549, rpn4 t-2-hex-H2 n=329. (B) t-2-hex activated protein aggregation was no longer observed after inhibition of protein synthesis with cycloheximide (CHX). Lower panel: Quantification of t-2-hex induced protein aggregates across cell populations. Number of analyzed cells: t-2-hex n=584, CHX +t-2-hex n=134. (C) Effect of pro-apoptotic t-2-hex on proteasomal activity. Proteasomal activity was quantified in whole cell extracts before (Cnt) or after treatment with 200 μM t-2-hex, t-2-hex-H2, or vehicle (DMSO) for 3 hr in wild-type or rpn4Δ cells (n=3). Activity of untreated wild-type cells was set to 100%. (D) Response of proteasomal subunit Rpn8 expression upon t-2-hex exposure. Rpn8 was expressed as a Tap fusion from its chromosomal locus and cells were treated or not with 200 μM t-2-hex for the indicated times. Rpn8 protein abundance was quantified by anti-Tap western blot (upper panel) and quantified relative to uninduced levels (n=2). (E) Proteasomal deficiency causes t-2-hex sensitivity. Growth of yeast wild-type and rpn4Δ cells was quantified upon the indicated t-2-hex concentrations (left panel) and the doubling time calculated (right panel), n=6. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0005 by Student´s unpaired t-test.
-
Figure 4—source data 1
PDF file containing the original uncropped western blots for Figure 4D, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93621/elife-93621-fig4-data1-v1.pdf
-
Figure 4—source data 2
Original files for western blot analysis displayed in Figure 4D.
- https://cdn.elifesciences.org/articles/93621/elife-93621-fig4-data2-v1.zip
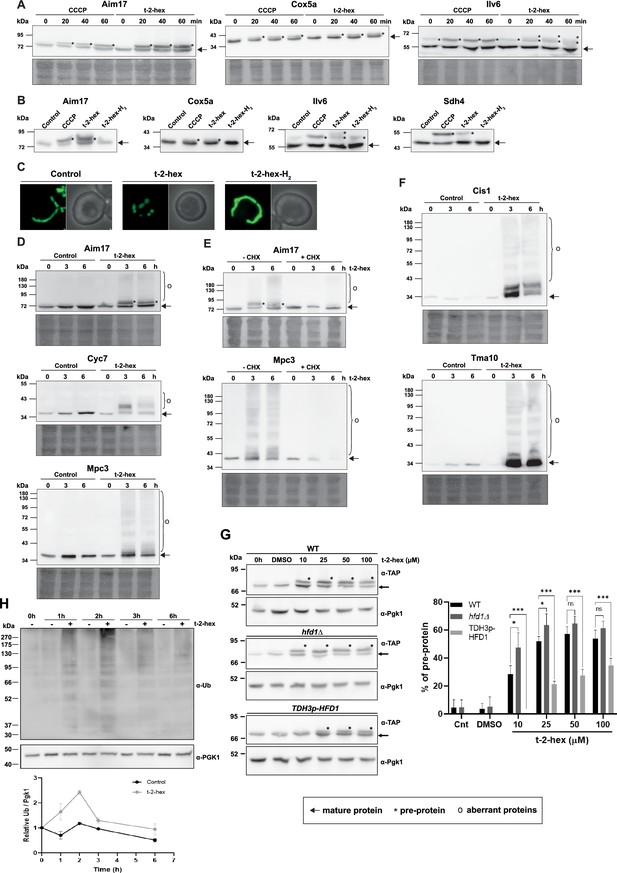
Trans-2-hexadecenal (t-2-hex) overload induces mitochondrial pre-protein accumulation and aggregation of de novo synthesized proteins.
(A) The appearance of unimported mitochondrial precursor proteins (*) was induced by the uncoupler CCCP (20 μM) or t-2-hex (200 μM) for the indicated times. Aim17, Cox5a, Ilv6, and Sdh4 were visualized in chromosomally Tap-tagged wild-type yeast strains by anti-Tap western blot. (B) Mitochondrial import block depends on t-2-hex unsaturation. The same strains as in (A) were treated with DMSO (control), CCCP (20 μM), t-2-hex (200 μM), or t-2-hex-H2 (200 μM) for 40 min and mitochondrial pre-protein accumulation was visualized by anti-Tap western blot. (C) Mitochondrial fragmentation depends on t-2-hex unsaturation. Yeast wild-type cells expressing mt-GFP were treated with vehicle (control), 200 μM t-2-hex or t-2-hex-H2 for 1 hr. (D) t-2-hex induces the formation of aberrant mitochondrial proteins. Yeast wild-type strains expressing Aim17-, Cyc7-, or Mpc3-Tap tagged fusion proteins from their chromosomal locus were treated with 200 μM t-2-hex for the indicated times. Fusion proteins were detected by anti-Tap western blot. (E) Inhibition of de novo protein synthesis abolishes the formation of aberrant mitochondrial proteins. Experimental conditions as in (D), but including a pretreatment with cycloheximide (CHX, 250 μg/ml) where indicated. (F) t-2-hex induces aberrant forms of highly expressed proteins. Yeast wild-type strains expressing Cis1- or Tma10-Tap tagged fusion proteins from their chromosomal locus were treated or not with 200 μM t-2-hex for the indicated times. Fusion proteins were detected by anti-Tap western blot. (G) Hfd1 gene dose determines the extent of t-2-hex-induced pre-protein accumulation. Aim17 pre-protein accumulation in response to different t-2-hex concentrations was quantified in wild-type, hfd1Δ and HFD1 overexpressing cells. Cells were either untreated, or treated with the indicated lipid concentrations or DMSO for 20 min. The percentage of pre-protein relative to total Aim17 protein was calculated in the right panel (n=4) (H) t-2-hex causes rapid protein ubiquitination. Yeast wild-type cells were treated for the indicated times with t-2-hex (200 μM) and protein ubiquitination visualized by anti-Ub western blot (upper panel) and quantified relative to the Pgk1 loading control (lower panel) (n=2).
-
Figure 5—source data 1
PDF file containing the original uncropped western blots for Figure 5, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93621/elife-93621-fig5-data1-v1.pdf
-
Figure 5—source data 2
Original files for western blot analysis displayed in Figure 5A-D.
- https://cdn.elifesciences.org/articles/93621/elife-93621-fig5-data2-v1.zip
-
Figure 5—source data 3
Original files for western blot analysis displayed in Figure 5E-H.
- https://cdn.elifesciences.org/articles/93621/elife-93621-fig5-data3-v1.zip
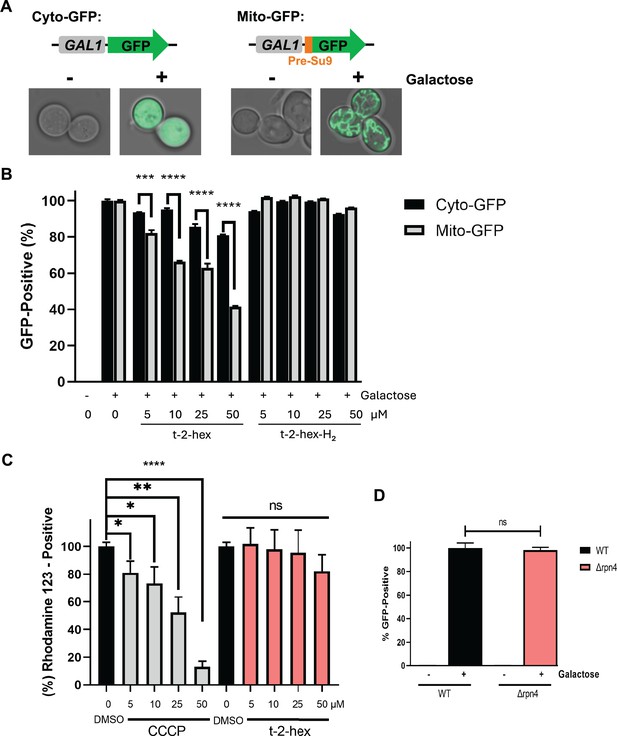
Quantification of the inhibition of mitochondrial protein import by cytometric mt-GFP measurements.
(A) Galactose-inducible expression of cytosolic or mitochondrial GFP from centromeric plasmids. Mitochondrial GFP contains a cysteine-free 69aa matrix targeting pre-sequence (Pre-Su9). Intracellular localization of both GFP markers before (-) and after (+) 2 hr of galactose induction. (B) Cytometric determination of mitochondrial and cytosolic GFP activity in the presence of the indicated t-2-hex and t-2-hex-H2 concentrations in yeast wild-type cells. (C) Modulation of mitochondrial membrane potential was determined by cytometric rhodamine123 measurements in yeast wild-type cells. The indicated t-2-hex and CCCP (mitochondrial uncoupler control) were applied for 2 hr. (D) Effect of impaired proteasomal activity on mt-GFP import. Mitochondrial GFP activity was determined cytometrically in live wild-type or Δrpn4 mutant cells. (n=3). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0005 by Student´s unpaired t-test, ns = not significant.
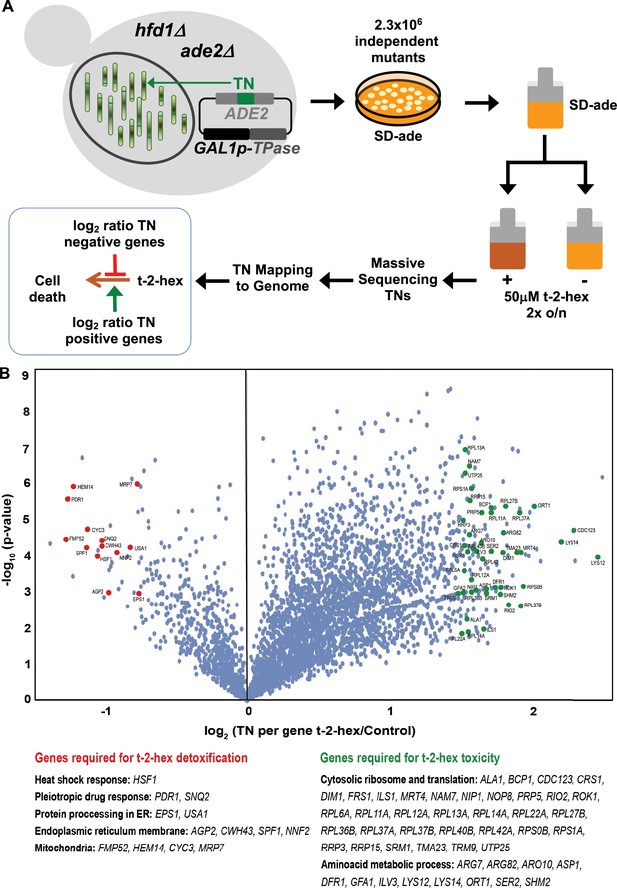
Functional genomics screen SATAY for the identification of pro- and anti-apoptotic functions upon trans-2-hexadecenal (t-2-hex) stress.
(A) Schematic outline of the SATAY experiment. hfd1Δ cells harboring the galactose-inducible transposase (TPase) and a transposon (TN) disrupting the ADE2 gene were grown in galactose-containing medium to induce transposition. TN-generated mutant cells were inoculated in synthetic glucose medium lacking adenine in the presence or absence of 50 μM t-2-hex, grown for several generations, and harvested for DNA extraction and sequencing of transposon insertion sites (TNs). TNs are mapped to the genome to identify genes that become required for proliferation under t-2-hex stress conditions (red) or genes whose mutation is beneficial for t-2-hex tolerance (green). (B) Identification of anti- and pro-apoptotic gene functions upon t-2-hex overload. Volcano plot showing the fold change of number of transposon insertions (TN) per gene of libraries grown in t-2-hex excess versus control conditions. TN under-enriched (anti-apoptotic) genes were analyzed with a log2 ratio below –0.75 and are summarized in Supplementary file 4. TN-enriched (pro-apoptotic) genes were analyzed with a log2 ratio >1.5 and are available in Supplementary file 5.
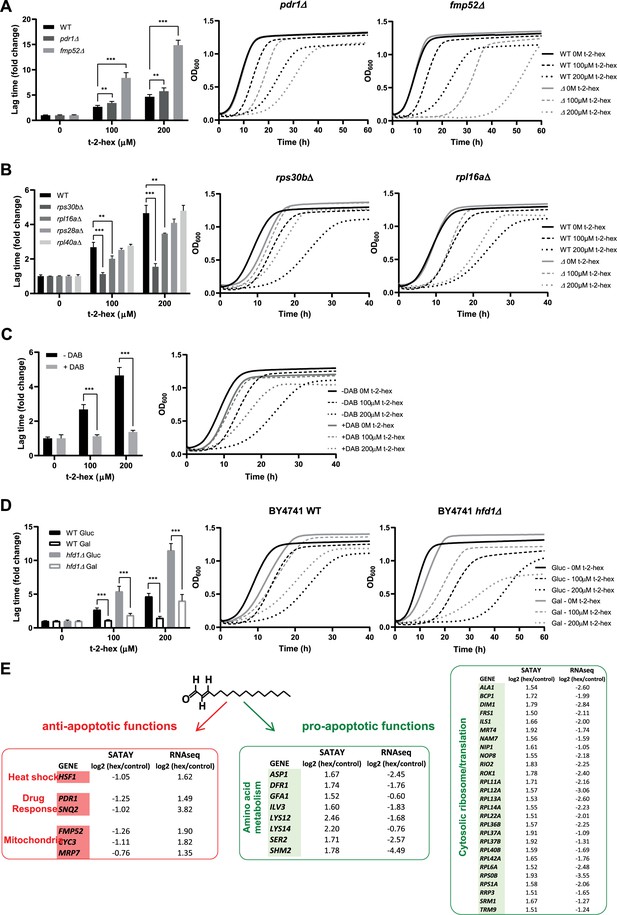
Functional analysis of pro- and anti-apoptotic genes upon trans-2-hexadecenal (t-2-hex) excess.
(A) The pleiotropic drug response activator Pdr1 and the mitochondrial protein Fmp52 are necessary for t-2-hex tolerance. Quantitative growth assay of wild-type, pdr1Δ, and fmp52Δ cells upon the indicated concentrations of t-2-hex. The relative lag time was quantified as an estimator of growth inhibition (n=3). (B) Deletion of ribosomal protein subunits causes t-2-hex resistance. As in (A), but using the yeast deletion strains rps30bΔ, rpl6aΔ, rps28aΔ, and rpl40aΔ. Growth curves are depicted only for the t-2-hex tolerant strains rps30bΔ and rpl6aΔ (n=3). (C) Inhibition of ribosomal biogenesis is beneficial for t-2-hex tolerance. Yeast wild-type cells were grown upon the indicated t-2-hex concentrations in the presence or absence of the inhibitor diazaborine (DAB, 20 μg/ml) (n=3). (D) Galactose growth improves t-2-hex tolerance. Growth of wild-type and hfd1Δ cells upon the indicated concentrations of t-2-hex on synthetic glucose or galactose media (n=3). **p<0.01, ***p<0.001 by Student´s unpaired t-test. (E) Summary of anti-apoptotic genes identified by SATAY with significant transcriptional up-regulation (red) and pro-apoptotic genes identified by SATAY with significant transcriptional down-regulation (green) according to our RNA-seq study.

t-2-hex targets the mitochondrial TOM complex.
(A) Hfd1 co-purifies with Tom70. Constitutively Hfd1-HA expressing cells were used in co-immunoprecipitation experiments from yeast whole cell extracts in the presence or not of endogenously expressed Tom22- or Tom70-Tap. Lower panel: Quantification of Hfd1 co-purification with respect to Tom20 and Tom70; n=6. (B) A chemoproteomic screen with t-2-hex alkyne identifies the TOM and Tim23 complexes as lipidation targets. Purified mitochondrial fractions from yeast wild-type cells were treated or not with the clickable analogue t-2-hex-Alkyne at high and low doses (100 μM or 10 μM). After addition of biotin to the modified proteins and purification with Streptavidin agarose pull-down, the protein identities of the trans-2-hexadecenal (t-2-hex) targets were determined by mass spectrometry. Cysteine-containing subunits of the TOM and Tim23 complexes are depicted as direct lipidation targets. The tables show the mean spectral reads for selected subunits in the chemoproteomic analysis for mock-treated (Ctrl) and t-2-hex treated mitochondria (n=2), as well as the log2 fold enrichment and adjusted p-value. (C) Tom40 is lipidated in vitro by t-2-hex. Upper panel: t-2-hex-Alkyne was used in the indicated concentrations to lipidate proteins in enriched mitochondrial preparations from Tom40-HA expressing cells or control cells. After t-2-hex addition, input samples were generated directly, while pull-down samples were treated with click chemistry for covalent biotin linkage and subsequent Streptavidin purification. Tom40-HA was detected in all samples by anti-HA western blot. Lower panel: Competition of Tom40 t-2-hex lipidation by free thiol groups. t-2-hex-Alkyne was used at 100 μM to lipidate Tom40-HA in mitochondrial preparations in the presence or absence of 2 mM DTT. (D) Mutations of TOM accessory subunits 20 or 70 cause t-2-hex tolerance. t-2-hex toxicity assays comparing wild-type cells with the cysteine-free Tom40 mutant (Tom40 CFREE) and deletion mutants tom20 or tom70. Lower panel: Quantitative comparison of growth performance (length of lag phase) of the tom20 and tom70 strains upon the indicated t-2-hex concentrations (n=3). **p<0.01, ***p<0.001 by Student´s unpaired t-test.
-
Figure 8—source data 1
PDF file containing the original uncropped western blots for Figure 8A and C, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93621/elife-93621-fig8-data1-v1.pdf
-
Figure 8—source data 2
Original files for western blot analysis displayed in Figure 8A and C.
- https://cdn.elifesciences.org/articles/93621/elife-93621-fig8-data2-v1.zip
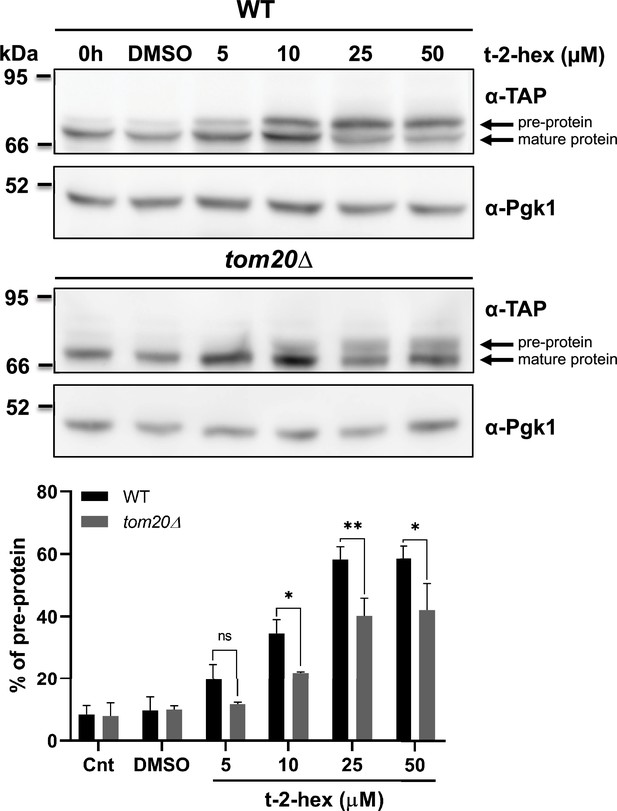
Tom20 function determines the extent of trans-2-hexadecenal (t-2-hex)-induced pre-protein accumulation.
Upper panels: Aim17 pre-protein accumulation in response to different t-2-hex concentrations was quantified in wild-type and tom20Δ cells. Cells were either untreated, or treated with the indicated lipid concentrations or DMSO for 20 min. The percentage of pre-protein relative to total Aim17 protein was calculated in the lower panel (n=3). *p<0.05, **p<0.01 by Student´s unpaired t-test, ns = not significant.
-
Figure 8—figure supplement 1—source data 1
PDF file containing the original uncropped western blots for Figure 8—figure supplement 1, indicating the relevant bands.
- https://cdn.elifesciences.org/articles/93621/elife-93621-fig8-figsupp1-data1-v1.pdf
-
Figure 8—figure supplement 1—source data 2
Original files for western blot analysis displayed in Figure 8—figure supplement 1.
- https://cdn.elifesciences.org/articles/93621/elife-93621-fig8-figsupp1-data2-v1.zip
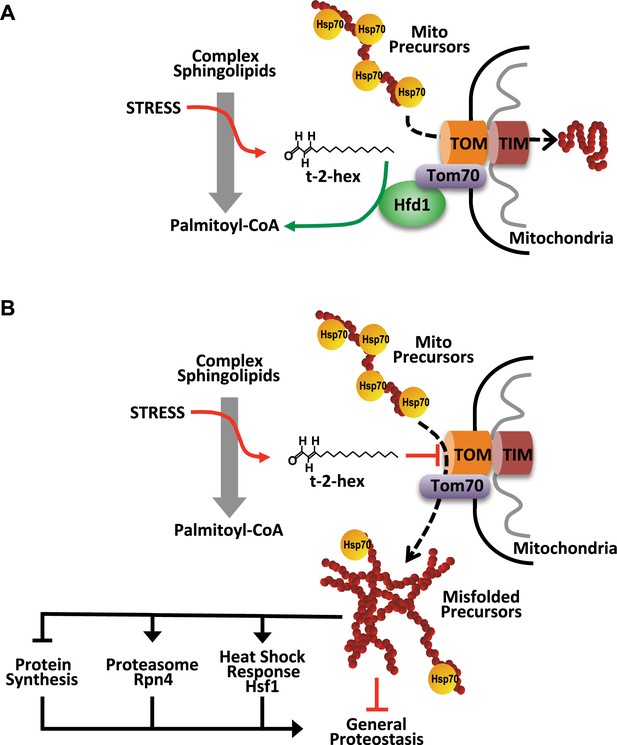
Model of the pro-apoptotic function of trans-2-hexadecenal (t-2-hex) and the anti-apoptotic function of Hfd1 at the mitochondrial Tom complex.
Upper panel: Under normal conditions, the Hfd1 lipid aldehyde dehydrogenase located at the Tom complex safeguards mitochondrial protein import by t-2-hex degradation. Lower panel: Upon severe stress conditions or in the absence of Hfd1 function, an excess of t-2-hex directly lipidates Tom subunits such as Tom40 and thus inhibits mitochondrial pre-protein transport across the outer mitochondrial membrane. The resulting proteostatic imbalance in the cytosol, if not sufficiently repaired or counteracted by the heat shock response, proteasomal clearance or diminished de novo protein synthesis, can induce apoptotic cell death.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Saccharomyces cerevisiae) | BY4741 | EUROSCARF | Genotype: MATa; his3∆1; leu2∆0; met15∆0; ura3∆0 | |
| Strain, strain background (Saccharomyces cerevisiae) | hfd1∆ | EUROSCARF | Genotype: BY4741 with hfd1Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | TDH3p-HFD1 | This study | Genotype: BY4741 with KanMX::pTDH3-HFD1 | |
| Strain, strain background (Saccharomyces cerevisiae) | rpn4∆ | Sergi Puig collection | Genotype: BY4741 with rpn4Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | WT Hsp104-GFP | Jacobson et al., 2012 | Genotype: BY4741 with HSP104-GFP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | rpn4∆ Hsp104-GFP | Jacobson et al., 2012 | Genotype: rpn4∆ with HSP104-GFP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | Rpn8-TAP | Ghaemmaghami et al., 2003 | Genotype: BY4741 with RPN8-TAP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | Aim17-TAP | Ghaemmaghami et al., 2003 | Genotype: BY4741 with AIM17-TAP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | Aim17-TAP hfd1∆ | This study | Genotype: Aim17-TAP with hfd1Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | Aim17-TAP TDH3p-HFD1 | This study | Genotype: Aim17-TAP with KanMX::pTDH3-HFD1 | |
| Strain, strain background (Saccharomyces cerevisiae) | Cox5a-TAP | Ghaemmaghami et al., 2003 | Genotype: BY4741 with COX5A-TAP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | Ilv6-TAP | Ghaemmaghami et al., 2003 | Genotype: BY4741 with ILV6-TAP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | Sdh4-TAP | Ghaemmaghami et al., 2003 | Genotype: BY4741 with SDH4-TAP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | Mpc3-TAP | Ghaemmaghami et al., 2003 | Genotype: BY4741 with MPC3-TAP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | Cyc7-TAP | Ghaemmaghami et al., 2003 | Genotype: BY4741 with CYC7-TAP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | Cis1-TAP | Ghaemmaghami et al., 2003 | Genotype: BY4741 with CIS1-TAP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | Tma10-TAP | Ghaemmaghami et al., 2003 | Genotype: BY4741 with TMA10-TAP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | ByK352 | Michel et al., 2017 | Genotype: BY4741 with ade2∆::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | ByK352 hfd1∆ | This study | Genotype: ByK352 with hfd1Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | pdr1∆ | Susana Rodríguez collection | Genotype: BY4741 with pdr1Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | fmp52∆ | Susana Rodríguez collection | Genotype: BY4741 with fmp52Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | rps30b∆ | Susana Rodríguez collection | Genotype: BY4741 with rps30bΔ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | rpl16a∆ | Susana Rodríguez collection | Genotype: BY4741 with rpl16aΔ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | rps28a∆ | Susana Rodríguez collection | Genotype: BY4741 with rps28aΔ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | rpl40a∆ | Susana Rodríguez collection | Genotype: BY4741 with rpl40aΔ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | Tom22-TAP | This study | Genotype: BY4741 with TOM22-TAP::HIS3 (Ghaemmaghami et al., 2003) and hfd1Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | Tom70-TAP | This study | Genotype: BY4741 with TOM70-TAP::HIS3 (Ghaemmaghami et al., 2003) and hfd1Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | Tom40-HA | Becker et al., 2011 | Genotype: YPH499 with tom40∆::ADE2 and plasmid pFL39-TOM40-HA | |
| Strain, strain background (Saccharomyces cerevisiae) | Tom40 WT | Qiu et al., 2013 | Genotype: YPH499 with tom40∆::ADE2 and plasmid pFL39-TOM40-TRP1 | |
| Strain, strain background (Saccharomyces cerevisiae) | Tom40 CFREE | Qiu et al., 2013 | Genotype: YPH499 with tom40∆::ADE2 and plasmid pFL39-TOM40CFREE-TRP1 | |
| Strain, strain background (Saccharomyces cerevisiae) | tom20∆ | This study | Genotype: BY4741 with tom20Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | tom70∆ | This study | Genotype: BY4741 with tom70Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | Aim17-TAP tom20∆ | This study | Genotype: Aim17-TAP with tom20Δ::KanMX | |
| Strain, strain background (Saccharomyces cerevisiae) | HSP104-GFP | Markus Tamas | Genotype: BY4741 with HSP104-GFP::HIS3 | |
| Strain, strain background (Saccharomyces cerevisiae) | HSP104-GFP rpn4∆ | Markus Tamas | Genotype: BY4741 with HSP104-GFP::HIS3 rpn4::KanMX | |
| Recombinant DNA reagent (plasmid) | pAG413- CYC1∆-lucCP+ | Rienzo et al., 2012 | AmpR, CEN, luciferase, HIS3 Destination vector for promoter cloning | |
| Recombinant DNA reagent (plasmid) | pUG6 | Güldener et al., 1996 | AmpR Generation of loxP–KanMX–loxP gene disruption cassettes | |
| Recombinant DNA reagent (plasmid) | pUG6-pTDH3 | This study | AmpR Generation of targeted KanMx::pTDH3 insertion cassettes | |
| Recombinant DNA reagent (plasmid) | pAG413- HFD1-lucCP+ | This study | pAG413-lucCP+ with HFD1 promoter | |
| Recombinant DNA reagent (plasmid) | pAG413- GRE2-lucCP+ | Rienzo et al., 2012 | pAG413-lucCP+ with GRE2 promoter | |
| Recombinant DNA reagent (plasmid) | pAG413- 4xHSE-lucCP+ | This study | pAG413-lucCP+ with 4 repetitions of HSE element | |
| Recombinant DNA reagent (plasmid) | pAG413-3xPACE-lucCP | This study | pAG413-lucCP+ with 3 repetitions of PACE element | |
| Recombinant DNA reagent (plasmid) | pAG413-3xPDRE-lucCP+ | Vanacloig-Pedros et al., 2019 | pAG413-lucCP+ with 3 repetitions of PDRE element | |
| Recombinant DNA reagent (plasmid) | pAG413- 3xAP-1-lucCP+ | Dolz-Edo et al., 2013 | pAG413-lucCP+ with 3 repetitions of AP-1 element | |
| Recombinant DNA reagent (plasmid) | pAG413- 3xCRE-lucCP+ | Dolz-Edo et al., 2013 | pAG413-lucCP+ with 3 repetitions of CRE element | |
| Recombinant DNA reagent (plasmid) | pBK549 | Michel and Kornmann, 2022 | AmpR, CEN, Ac transposase under GAL1 promoter, ADE2 gene interrupted by the MiniDs transposon, URA3 | |
| Recombinant DNA reagent (plasmid) | pVT100-mtGFP | Westermann and Neupert, 2000 | AmpR, 2 µ, ADH1 promoter, mitochondria-targeted GFP, URA3 | |
| Recombinant DNA reagent (plasmid) | pAG426- GPD-HFD1-HA | Markus Poft collection | AmpR, 2 µ, GPD promoter, HFD1 CDS with C-term HA tag, URA3 | |
| Recombinant DNA reagent (plasmid) | pFL39- TOM40-HA | Becker et al., 2011 | AmpR, CEN, TOM40 promoter, TOM40 CDS with C-term HA tag, TRP1 | |
| Recombinant DNA reagent (plasmid) | pFL39-TOM40WT | Qiu et al., 2013 | AmpR, CEN, TOM40 promoter, Tom40 WT, TRP1 | |
| Recombinant DNA reagent (plasmid) | pFL39-TOM40CFREE | Qiu et al., 2013 | AmpR, CEN, TOM40 promoter, Tom40 with mutated cysteines, TRP1 | |
| Recombinant DNA reagent (plasmid) | pRS416-GFP | Fita-Torró et al., 2023 | AmpR, CEN, GAL1 promoter, GFP, URA3 | |
| Recombinant DNA reagent (plasmid) | pYX113-mtGFP | Westermann and Neupert, 2000 | AmpR, CEN, GAL1 promoter, Pre-Su9-GFP, URA3 | |
| Sequence-based reagent (cloning primer) | BspEI-3PACE-1 | This study | Cloning PACE sites into pAG413-CYC1∆-lucCP+ | Sequence 5’ ->3’: P-CCGGCGGTGGCAAAGATATCGGTGGCAAAGTAATCGGTGGCAAAT |
| Sequence-based reagent (cloning primer) | BspEI-3PACE-2 | This study | Sequence 5’ ->3’: P-CCGGATTTGCCACCGATTACTTTGCCACCGATATCTTTGCCACCG | |
| Sequence-based reagent (cloning primer) | BspEI-4HSE-1 | This study | Cloning HSE sites into pAG413-CYC1∆-lucCP+ | Sequence 5’ ->3’: P-CCGGCGATATCTTCTAGAAGCTTCTAGAAGT |
| Sequence-based reagent (cloning primer) | BspEI-4HSE-2 | This study | Sequence 5’ ->3’: P-CCGGACTTCTAGAAGCTTCTAGAAGATATCG | |
| Sequence-based reagent (PCR primer) | HFD1-995SacI | This study | Cloning HFD1 promoter from BY4741 into pAG413-CYC1∆-lucCP+ | Sequence 5’ ->3’: GCCGAGCTCTGATGACAGTAATAACCAACTCG |
| Sequence-based reagent (PCR primer) | HFD1-1SmaI | This study | Sequence 5’ ->3’: TCCCCCGGGGTTGGTGATAAATTACTATGGCTATGGTTT | |
| Sequence-based reagent (PCR primer) | TDH3pEcoRVFw | This study | Cloning TDH3 promoter from BY4741 into pUG6 | Sequence 5’ ->3’: GGCCGATATCATTATCAATACTGCCATTTC |
| Sequence-based reagent (PCR primer) | TDH3pSpeIRev | This study | Sequence 5’ ->3’: GGCCACTAGTTTGTTTGTTTATGTGTGTTTATTCG | |
| Sequence-based reagent (PCR primer) | HFD1-Del-1 | This study | Amplification of KanMX disruption cassette from pUG6 for hfd1∆ strain generation | Sequence 5’ ->3’: AAAAGGAATATTCTAAAACCATAGCCATAGTAATTTATCACCAACCAGCTGAAGCTTCGTACGC |
| Sequence-based reagent (PCR primer) | HFD1-Del-2 | This study | Sequence 5’ ->3’: AGGTTACTTATACATCAAATAATTAATTAACCTTAAACATTACGTGCATAGGCCACTAGTGGATCTG | |
| Sequence-based reagent (PCR primer) | HFD1-TDH3-1 | This study | Amplification of KanMX::pTDH3 cassette from pUG6-pTDH3 for HFD1 promoter replacement | Sequence 5’ ->3’: CAACGAATTTCCAGCCAAAAATTCCGAGTAGTTCATGATGAAAGAGCTGAAGCTTCGTACGCTGC |
| Sequence-based reagent (PCR primer) | HFD1-TDH3-2 | This study | Sequence 5’ ->3’: AGACACTGGGGTATAATTCAATATTTTTGAGCCGTCGTTTGACATTTTGTTTGTTTATGTGTGTTTATTCG | |
| Sequence-based reagent (PCR primer) | TOM20-Del-1 | This study | Amplification of KanMX disruption cassette from pUG6 for tom20∆ strain generation | Sequence 5’ ->3’: GAAACATTGCCTCAAGTGCCACCTTCATAAAGTTTATTTTCTATTCAGCTGAAGCTTCGTACGC |
| Sequence-based reagent (PCR primer) | TOM20-Del-2 | This study | Sequence 5’ ->3’: GTAAAAGAAACAAAAACGGAGAAAAAAAGCAAGCAAAATGTTACTCGCATAGGCCACTAGTGGATCTG | |
| Sequence-based reagent (PCR primer) | TOM70-Del-1 | This study | Amplification of KanMX disruption cassette from pUG6 for tom70∆ strain generation | Sequence 5’ ->3’: GATTCGGAAGTGAAATTACAGCTCACATCTAGGTTCTCAATTGCCACAGCTGAAGCTTCGTACGC |
| Sequence-based reagent (PCR primer) | TOM70-Del-2 | This study | Sequence 5’ ->3’: TTAGTTTTTGTCTTCTCCTAAAAGTTTTTAAGTTTATGTTTACTGTGCATAGGCCACTAGTGGATCTG | |
| Sequence-based reagent (PCR primer) | P5_MiniDs | This study | Forward primer for all libraries | Sequence 5’ ->3’: AATGATACGGCGACCACCGAGATCTACtccgtcccgcaagttaaatatga |
| Sequence-based reagent (PCR primer) | P7_MiniDs_i1 | This study | Reverse primer for Control_Hin1II library | Sequence 5’ ->3’: CAAGCAGAAGACGGCATACGAGATCGAGTAATacgaaaacgaacgggataaatac |
| Sequence-based reagent (PCR primer) | P7_MiniDs_i2 | This study | Reverse primer for Control_MboI library | Sequence 5’ ->3’: CAAGCAGAAGACGGCATACGAGATTCTCCGGAacgaaaacgaacgggataaatac |
| Sequence-based reagent (PCR primer) | P7_MiniDs_i3 | This study | Reverse primer for t-2-hex_Hin1II library | Sequence 5’ ->3’: CAAGCAGAAGACGGCATACGAGATAATGAGCGacgaaaacgaacgggataaatac |
| Sequence-based reagent (PCR primer) | P7_MiniDs_i4 | This study | Reverse primer for t-2-hex_MboI library | Sequence 5’ ->3’: CAAGCAGAAGACGGCATACGAGATGGAATCTCacgaaaacgaacgggataaatac |
| Sequence-based reagent (PCR primer) | 688_minids SEQ1210 | This study | Sequencing primer | Sequence 5’ ->3’: TTTACCGACCGTTACCGACCGTTTTCATCCCTA |
| Sequence-based reagent (PCR primer) | Custom_ index1 | This study | Index identification | Sequence 5’ ->3’: GGTTTTCGATTACCGTATTTATCCCGTTCGTTTTCGT |
| Antibody | α-TAP (TAP Tag Rabbit Polyclonal) | Invitrogen | Product # CAB1001 | WB (1:5000) |
| Antibody | α-HA (HA Tag Mouse Monoclonal) | Proteintech | Cat No. 66006–2-Ig | WB (1:10000) |
| Antibody | α-Ubi (Ubiquitin P4D1 Mouse Monoclonal) | Santa Cruz Biotechnology | Cat No. SC-8017 | WB (1:1000) |
| Antibody | α-Pgk1 (PGK1 Mouse Monoclonal (22C5D8)) | Invitrogen | Product # 10073994 | WB (1:5000) |
| Antibody | α-rabbit (anti-rabbit IgG HRP) | Cytiva | Cat No. NA934 | WB (1:10000) |
| Antibody | α-mouse (anti-mouse IgG HRP) | Cytiva | Cat No. NA931 | WB (1:10000) |
| Chemical compound, drug | trans-2-hexadecenal (t-2-hex) | Avanti Research | 857459 P | |
| Chemical compound, drug | Hexadecanal (t-2-hex-H2) | Avanti Research | 857458 M | |
| Chemical compound, drug | t-2-hex-Alkyne (E)–2-Hexadecenal Alkyne | Cayman Chemical | Item No. 20714 | |
| Chemical compound, drug | Diazaborine (DAB) (2-(5-Bromo-2-thienyl)–2,3-dihydro-1H-naphtho[1,8-de][1,3,2]diazaborine) | TCI Chemicals | Cat No. B3868 | 20 μg/mL |
| Chemical compound, drug | D-Luciferin free acid, 99% | Synchem | Product No. S039 | 0.5 mM |
| Chemical compound, drug | Suc-LLVY-AMC | Bachem | Product No. 4011369 | 0.1 mM |
Additional files
-
Supplementary file 1
Transcriptomic analysis (RNA seq) of t-2-hex-induced gene functions.
- https://cdn.elifesciences.org/articles/93621/elife-93621-supp1-v1.xlsx
-
Supplementary file 2
Transcriptomic analysis (RNA seq) of t-2-hex repressed gene functions.
- https://cdn.elifesciences.org/articles/93621/elife-93621-supp2-v1.xlsx
-
Supplementary file 3
Comparison of the transcriptomic response to mitochondrial import stress and t-2-hex overload.
- https://cdn.elifesciences.org/articles/93621/elife-93621-supp3-v1.xlsx
-
Supplementary file 4
SATAY underenriched gene functions upon t-2-hex stress.
- https://cdn.elifesciences.org/articles/93621/elife-93621-supp4-v1.xlsx
-
Supplementary file 5
SATAY enriched gene functions upon t-2-hex stress.
- https://cdn.elifesciences.org/articles/93621/elife-93621-supp5-v1.xlsx
-
Supplementary file 6
Results of the chemoproteomic protein identification comparing t-2-hex-alkyne and solvent-treated cell extracts at 10 μM and 100 μM.
- https://cdn.elifesciences.org/articles/93621/elife-93621-supp6-v1.xlsx
-
Supplementary file 7
Yeast strains used in this study.
- https://cdn.elifesciences.org/articles/93621/elife-93621-supp7-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/93621/elife-93621-mdarchecklist1-v1.docx





