Transcription: Watching single molecules in action
Single-molecule techniques allow researchers to follow, in real-time, the details of biochemical reactions that are obscured when experiments are carried out on a large number of molecules (Kapanidis and Strick, 2009). These details include transient molecular species and the kinetics of molecular complexes. For example, the ability to observe both the assembly and disassembly of molecular complexes, rather than the equilibrium between these two process, could provide mechanistic insights into the way that these complexes perform various functions in cells. Moreover, if one of the molecules involved in a biochemical reaction can be tethered to a solid support, such as a glass coverslip, and placed in a solution, the effects of random diffusion can also be eliminated: this will make it possible to continuously monitor how the tethered molecule interacts with other molecules in solution.
The future of the field lies in techniques that can directly and simultaneously monitor an ever-expanding number of reagents with ever-increasing throughput and ever-better time resolution (Friedman et al., 2006; Uemura et al., 2010; Hoskins et al., 2011; Revyakin et al., 2012; Chen et al., 2014). Now, in eLife, Zhengjian Zhang, Andrey Revyakin and co-workers have applied these principles to capture the full reaction cycle of a simple yet highly kinetic system, the T7 RNA polymerase (RNAP), in unprecedented detail (Zhang et al., 2014). T7 RNAP is an enzyme that binds to a stretch of DNA called the T7 promoter and starts the process by which downstream genes are transcribed to produce RNA molecules (which are subsequently translated to produce proteins).
Although the multi-step process of transcription has been studied extensively, the fast kinetics of T7 RNAP present unique challenges, and these have made it difficult to study the workings of this enzyme at single-molecule resolution (Skinner et al., 2004; Thomen et al., 2008; Skinner et al., 2011). Indeed, T7 RNAP is capable of initiating transcription and escaping from the promoter (an important part of the transcription process) within a second of binding to the promoter. It can also transcribe several hundreds of nucleotides per second. Therefore, in order to assess the full transcription cycle of T7 RNAP, it is essential to rapidly and directly monitor two processes: the binding of RNAP to DNA, and the synthesis of RNA. Until now, however, techniques for the direct detection of the nascent RNA have been too slow to follow the fast kinetics of T7 RNAP.
Zhang, Revyakin and co-workers—who are based at the Janelia Farm Research Campus and the University of California Berkeley—developed a method that allows them to directly and independently monitor these two processes. The binding of RNAP to DNA was monitored by adding a fluorescent label (a Cy5 molecule) to the RNAP, and the nascent RNA was monitored with a fluorescent DNA probe that consists of a single strand of DNA attached to a fluorescent Cy3 molecule (Figure 1). The technique used to probe for the nascent RNA is called fastFISH because it is a faster version of an existing technique called fluorescence in situ hybridization (FISH).
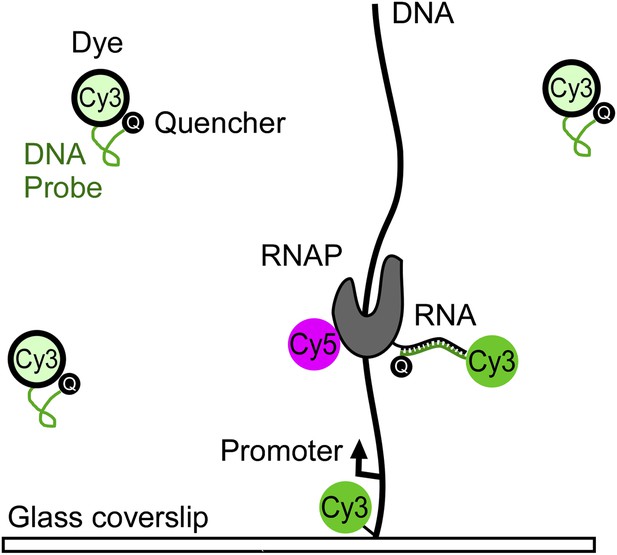
Tracking the transcription of a DNA molecule with the fastFISH method.
The DNA molecule is labelled with a fluorescent Cy3 molecule and tethered to a glass cover slip; an optical field (not shown) is then used to generate fluorescence, which allows the position of the DNA molecule to be determined. Next, the Cy3 label on the DNA molecule is bleached out so Cy3 can be used to label the DNA probe. The T7 RNA polymerase (RNAP), which is labelled with a fluorescent Cy5 molecule, starts to transcribe the DNA molecule to produce an RNA molecule, and the Cy3 molecule in the DNA probe starts to fluoresce when the probe binds to the nascent RNA molecule. The RNAP and the DNA probe can be monitored independently and simultaneously (with a time resolution of 80 milliseconds) because the Cy5 and Cy3 molecules fluoresce at different wavelengths. It is also possible to distinguish between T7 RNAP bound to a promoter on the DNA and T7 RNAP transcribing the DNA because, in the latter case, the fluorescence from the Cy5 will coincide with fluorescence from the Cy3.
FastFISH is based on two key ideas: first, in order to achieve high rates of hybridization between the DNA probe and the nascent RNA, the probe must not form stable secondary structures. This is achieved by using only three of the four nucleotides found in DNA—adenine, thymine and guanine—to design the DNA probe, and results in hybridization rates that are close to the maximum possible rate allowed by diffusion. However, relatively high concentrations of the DNA probe are needed to achieve a time resolution of better than 1 second, and this causes a problem: the high levels of background fluorescence from the DNA probes will overwhelm the fluorescence from the single molecules we want to study. Zhang et al. overcome this problem with a second key idea—they attach a quencher molecule to their DNA probe to switch off the fluorescence of the Cy3 molecule. However, the fluorescence is switched on when the DNA probes binds to the nascent RNA. By eliminating background fluorescence, these ‘self-quenching’ DNA probes are able to detect nascent RNA with a time resolution on the order of 80 milliseconds.
Using this approach, Zhang et al. measured the key kinetic features of the T7 RNAP transcription cycle. The binding of T7 RNAP to promoter DNA is very fast, limited only by diffusion, but this binding does not always result in transcription because the RNAP can also dissociate from the DNA. However T7 RNAP is biased towards productive transcription, and is capable of initiating transcription and carrying out promoter escape within 0.2 seconds of binding to the promoter.
By demonstrating the ability to directly monitor multiple components of the viral T7 transcription machinery in real time with a resolution of better than one second, Zhang, Revyakin and co-workers show how single-molecule experimentation is rapidly evolving into multi-molecule experimentation. This occurs at a key moment in biochemistry: ever-larger macromolecular complexes can routinely be purified and analysed structurally (Gonen et al., 2012), but performing detailed kinetic analysis on these complexes remains challenging. By developing ways to monitor multiple components simultaneously in real-time, and by providing ways to work in near-physiological conditions (Hoskins et al., 2011), single-molecule experimentation will continue to provide new tools to help narrow the gap between biochemistry carried out in vitro and in vivo.
References
-
High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescenceProceedings of the National Academy of Sciences of the United States of America 111:664–669.https://doi.org/10.1073/pnas.1315735111
-
The structure of purified kinetochores reveals multiple microtubule-attachment sitesNature Structural & Molecular Biology 19:925–929.https://doi.org/10.1038/nsmb.2358
-
Biology, one molecule at a timeTrends in Biochemical Sciences 34:234–243.https://doi.org/10.1016/j.tibs.2009.01.008
-
Transcription initiation by human RNA polymerase II visualized at single-molecule resolutionGenes & Development 26:1691–1702.https://doi.org/10.1101/gad.194936.112
-
Promoter binding, initiation and elongation by bacteriophage T7 RNA polymerase. A single-molecule view of the transcription cycleJournal of Biological Chemistry 279:3239–3244.https://doi.org/10.1074/jbc.M310471200
-
Downstream DNA tension regulates the stability of the T7 RNA polymerase initiation complexBiophysical Journal 100:1034–1041.https://doi.org/10.1016/j.bpj.2010.11.092
-
T7 RNA polymerase studied by force measurements varying cofactor concentrationBiophysical Journal 95:2423–2433.https://doi.org/10.1529/biophysj.107.125096
Article and author information
Author details
Publication history
- Version of Record published: January 28, 2014 (version 1)
Copyright
© 2014, Monnet and Strick
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,287
- views
-
- 56
- downloads
-
- 0
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Biochemistry and Chemical Biology
Transporter research primarily relies on the canonical substrates of well-established transporters. This approach has limitations when studying transporters for the low-abundant micromolecules, such as micronutrients, and may not reveal physiological functions of the transporters. While d-serine, a trace enantiomer of serine in the circulation, was discovered as an emerging biomarker of kidney function, its transport mechanisms in the periphery remain unknown. Here, using a multi-hierarchical approach from body fluids to molecules, combining multi-omics, cell-free synthetic biochemistry, and ex vivo transport analyses, we have identified two types of renal d-serine transport systems. We revealed that the small amino acid transporter ASCT2 serves as a d-serine transporter previously uncharacterized in the kidney and discovered d-serine as a non-canonical substrate of the sodium-coupled monocarboxylate transporters (SMCTs). These two systems are physiologically complementary, but ASCT2 dominates the role in the pathological condition. Our findings not only shed light on renal d-serine transport, but also clarify the importance of non-canonical substrate transport. This study provides a framework for investigating multiple transport systems of various trace micromolecules under physiological conditions and in multifactorial diseases.
-
- Biochemistry and Chemical Biology
- Cell Biology
Mediator of ERBB2-driven Cell Motility 1 (MEMO1) is an evolutionary conserved protein implicated in many biological processes; however, its primary molecular function remains unknown. Importantly, MEMO1 is overexpressed in many types of cancer and was shown to modulate breast cancer metastasis through altered cell motility. To better understand the function of MEMO1 in cancer cells, we analyzed genetic interactions of MEMO1 using gene essentiality data from 1028 cancer cell lines and found multiple iron-related genes exhibiting genetic relationships with MEMO1. We experimentally confirmed several interactions between MEMO1 and iron-related proteins in living cells, most notably, transferrin receptor 2 (TFR2), mitoferrin-2 (SLC25A28), and the global iron response regulator IRP1 (ACO1). These interactions indicate that cells with high MEMO1 expression levels are hypersensitive to the disruptions in iron distribution. Our data also indicate that MEMO1 is involved in ferroptosis and is linked to iron supply to mitochondria. We have found that purified MEMO1 binds iron with high affinity under redox conditions mimicking intracellular environment and solved MEMO1 structures in complex with iron and copper. Our work reveals that the iron coordination mode in MEMO1 is very similar to that of iron-containing extradiol dioxygenases, which also display a similar structural fold. We conclude that MEMO1 is an iron-binding protein that modulates iron homeostasis in cancer cells.





