Phylogeny: Unravelling a can of worms
The written accounts and reasoning associated with how the species we currently use for biomedical research were selected as model systems have rarely included weighty consideration of either their evolutionary or natural histories (Alfred and Baldwin, 2015). Instead, organisms such as Drosophila, Caenorhabditis elegans, zebrafish and mice were primarily chosen as model organisms for purely practical reasons. For example, many model organisms produce large numbers of offspring, have features that make them easy to examine (such as transparent embryos), or are easy to domesticate and look after in the laboratory. Paradoxically, while this approach has been remarkably successful in advancing our understanding of life, it has also made us acutely aware of how much more biology we have yet to comprehend.
Some have argued that further research into the model organisms that dominate much of biomedical research today could fill in the many gaps that exist in our understanding of life, but these organisms give a biased and ultimately poor statistical representation of the ∼30 million species of animals that populate our planet (Brusca and Brusca, 2003). Our best chances of uncovering new biology and acquiring a truly transformative understanding of life are therefore to be found in the laboratory of nature. As technology advances and allows us to examine aspects of biology that were not previously accessible to scientific interrogation, we may want to reconsider how we choose animals as new model systems. Instead of selecting them purely on the biological attributes they conveniently exaggerate for our scientific interests, we could also consider the ecological and evolutionary histories that may have helped produce such attributes.
These considerations are particularly timely in view of current efforts to develop and characterize invertebrate model systems for studying regeneration and parasitism. Now, in eLife, Christopher Laumer of Harvard University, Andreas Hejnol of the University of Bergen and Gonzalo Giribet, also from Harvard, have unravelled the phylogenetic tree of the Platyhelminthes, which are more commonly known as the flatworms (Laumer et al., 2015).
Animals possessing bilateral symmetry are presently grouped into three main branches in the metazoan tree of life. The Deuterostomes (the evolutionary lineage to which humans belong) are represented by a number of model organisms including mice, fish, sea squirts, sea urchins and, of course, humans. The second branch, the Ecdysozoa, is presently represented in biomedical research by the fruit fly Drosophila melanogaster and the roundworm nematode C. elegans. However, the third branch, the Lophotrochozoa, remains among the most undersampled and understudied collection of complex organisms on the planet. This is despite the fact that it encompasses a collection of animals with an assortment of body plans, biological attributes, and ecological adaptations that is unmatched by the other two bilaterian metazoan branches combined. Within the Lophotrochozoa, no group of animals manifests these attributes as clearly as the Platyhelminthes: the diversity of body plans, developmental plasticity and ecological adaptations displayed by these flatworms is remarkable.
Laumer, Hejnol and Giribet report on a comprehensive analysis of the evolutionary relationships among the Platyhelminthes using a survey of genomes and transcriptomes that represents all free-living (i.e., non-parasitic) flatworm orders. This work is the first of its type for the Platyhelminthes and ultimately provides a modern hypothesis that should help us to understand how this extraordinarily diverse group of animals evolved.
By comparing hundreds of nuclear protein coding genes, Laumer et al. were able to derive a phylogeny with at least two important and intriguing attributes. Firstly, key evolutionary transitions within the Platyhelminthes unexpectedly featured the involvement of ‘microturbellarian’ (microflatworm) groups (Figure 1). Secondly, a novel scenario that explains the interrelationships between free-living and parasitic flatworms provides unique opportunities for shedding light on the origins and biological consequences of the parasitic lifestyle in these animals.
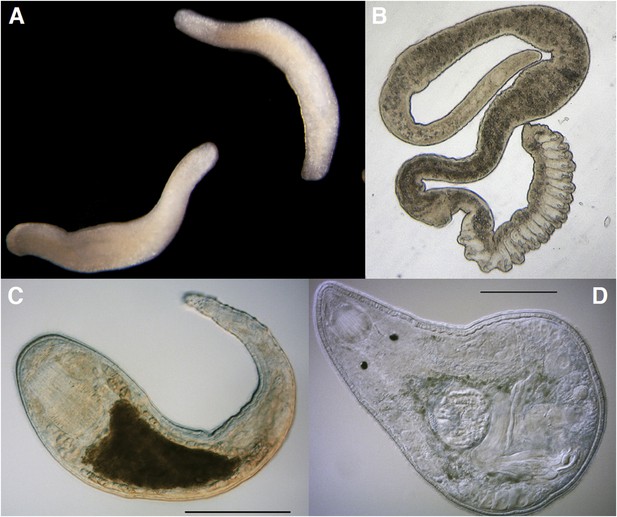
The Platyhelminthes show a diverse range of body plans and developmental characteristics.
(A) Two adult Bothrioplana semperi (Bothrioplanida) swimming in different directions. Adults are approximately 1.5 mm outstretched. Collected from near Saw Mill brook, Estabrook Woods, Concord, MA. (B) Adult of an undescribed species of Polystyliphora (Proseriata), collected from the interstices of marine sand in Bocas del Toro, Panama. Scale bar unavailable. (C) Adult specimen of an undescribed blind species of Microdalyellia (Rhabdocoela), collected in fresh water from Mt. Blue Spring, Wompatuck State Park, MA. (D) Adult, undetermined species of Nannorhynchides (Rhabdocoela), collected in brackish water at low tide near Sage Lot Pond outlet, Mashpee, MA. Scale bars: 100 µm. Samples in panels A, C and D photographed by Christopher E Laumer. Sample in panel B photographed by Marco Curini-Galletti.
The Tricladida order of flatworms contains the important model system Schmidtea mediterranea, which is used to study tissue regeneration and development. An intriguing point raised by the phylogenetic tree produced by Laumer et al. is that this order may be evolutionarily equidistant to two other orders (Prolecithophora and Fecampiida). This new relationship will have to be taken into account from this point onward when considering how the regenerative properties displayed by these three groups of animals evolved.
The work of Laumer et al. makes it clear that we should embrace an approach that involves morphological studies, evolutionary developmental biology and evolutionary genomics when selecting organisms for experimental interrogation. The evidence reported for the importance of microturbellarians (Figure 1) in the evolution of Platyhelminthes may ultimately prove to be the single most important contribution of the present body of work. Microturbellarians have not captured the attention of researchers like the best-known branches of the clearly much larger and phylogenetically diverse flatworms (e.g., planarians, polyclads, and neodermatans). It is my suspicion that this paper will bring an end to their relative obscurity.
References
Article and author information
Author details
Publication history
- Version of Record published: April 16, 2015 (version 1)
Copyright
© 2015, Sánchez Alvarado
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,410
- views
-
- 135
- downloads
-
- 2
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Genetics and Genomics
Telomeres, which are chromosomal end structures, play a crucial role in maintaining genome stability and integrity in eukaryotes. In the baker’s yeast Saccharomyces cerevisiae, the X- and Y’-elements are subtelomeric repetitive sequences found in all 32 and 17 telomeres, respectively. While the Y’-elements serve as a backup for telomere functions in cells lacking telomerase, the function of the X-elements remains unclear. This study utilized the S. cerevisiae strain SY12, which has three chromosomes and six telomeres, to investigate the role of X-elements (as well as Y’-elements) in telomere maintenance. Deletion of Y’-elements (SY12YΔ), X-elements (SY12XYΔ+Y), or both X- and Y’-elements (SY12XYΔ) did not impact the length of the terminal TG1-3 tracks or telomere silencing. However, inactivation of telomerase in SY12YΔ, SY12XYΔ+Y, and SY12XYΔ cells resulted in cellular senescence and the generation of survivors. These survivors either maintained their telomeres through homologous recombination-dependent TG1-3 track elongation or underwent microhomology-mediated intra-chromosomal end-to-end joining. Our findings indicate the non-essential role of subtelomeric X- and Y’-elements in telomere regulation in both telomerase-proficient and telomerase-null cells and suggest that these elements may represent remnants of S. cerevisiae genome evolution. Furthermore, strains with fewer or no subtelomeric elements exhibit more concise telomere structures and offer potential models for future studies in telomere biology.
-
- Genetics and Genomics
- Neuroscience
Genome-wide association studies have revealed >270 loci associated with schizophrenia risk, yet these genetic factors do not seem to be sufficient to fully explain the molecular determinants behind this psychiatric condition. Epigenetic marks such as post-translational histone modifications remain largely plastic during development and adulthood, allowing a dynamic impact of environmental factors, including antipsychotic medications, on access to genes and regulatory elements. However, few studies so far have profiled cell-specific genome-wide histone modifications in postmortem brain samples from schizophrenia subjects, or the effect of antipsychotic treatment on such epigenetic marks. Here, we conducted ChIP-seq analyses focusing on histone marks indicative of active enhancers (H3K27ac) and active promoters (H3K4me3), alongside RNA-seq, using frontal cortex samples from antipsychotic-free (AF) and antipsychotic-treated (AT) individuals with schizophrenia, as well as individually matched controls (n=58). Schizophrenia subjects exhibited thousands of neuronal and non-neuronal epigenetic differences at regions that included several susceptibility genetic loci, such as NRG1, DISC1, and DRD3. By analyzing the AF and AT cohorts separately, we identified schizophrenia-associated alterations in specific transcription factors, their regulatees, and epigenomic and transcriptomic features that were reversed by antipsychotic treatment; as well as those that represented a consequence of antipsychotic medication rather than a hallmark of schizophrenia in postmortem human brain samples. Notably, we also found that the effect of age on epigenomic landscapes was more pronounced in frontal cortex of AT-schizophrenics, as compared to AF-schizophrenics and controls. Together, these data provide important evidence of epigenetic alterations in the frontal cortex of individuals with schizophrenia, and remark for the first time on the impact of age and antipsychotic treatment on chromatin organization.





