Co-expression of Foxa.a, Foxd and Fgf9/16/20 defines a transient mesendoderm regulatory state in ascidian embryos
Abstract
In many bilaterian embryos, nuclear β-catenin (nβ-catenin) promotes mesendoderm over ectoderm lineages. Although this is likely to represent an evolutionary ancient developmental process, the regulatory architecture of nβ-catenin-induced mesendoderm remains elusive in the majority of animals. Here, we show that, in ascidian embryos, three nβ-catenin transcriptional targets, Foxa.a, Foxd and Fgf9/16/20, are each required for the correct initiation of both the mesoderm and endoderm gene regulatory networks. Conversely, these three factors are sufficient, in combination, to produce a mesendoderm ground state that can be further programmed into mesoderm or endoderm lineages. Importantly, we show that the combinatorial activity of these three factors is sufficient to reprogramme developing ectoderm cells to mesendoderm. We conclude that in ascidian embryos, the transient mesendoderm regulatory state is defined by co-expression of Foxa.a, Foxd and Fgf9/16/20.
https://doi.org/10.7554/eLife.14692.001Introduction
The mesoderm, endoderm and ectoderm arise during embryonic development by a process termed germ layer segregation. In many species, at least part of the endoderm and mesoderm derive from transient ‘mesendoderm’ precursors, as is the case in ascidian embryos (Kimelman and Griffin, 2000; Rodaway and Patient, 2001). However, the precise nature of this induced regulatory state is not well understood. In ascidians, the first animal-vegetal (A-V) oriented cell division generates the eight-cell stage embryo and segregates the mesendoderm and some neural lineages into two pairs of vegetal founder lineages (the A- and B-line) and the ectoderm (epidermis and neural) into two pairs of animal lineages (a- and b-line) (Conklin, 1905; Nishida, 1987). This study focuses on the A-line mesendoderm lineages. From the 8- to 16-cell stage, the two A4.1 blastomeres divide medio-laterally to generate the two pairs of neuro-mesendodermal NNE cells, for notochord/neural/endoderm (Figure 1a). NNE cells then divide along the A-V axis to generate NN cells (notochord/neural) and E cells (mostly endoderm) at the 32-cell stage (Figure 1a). Subsequently, NN cells segregate into notochord and neural lineages at the 64-cell stage. At this stage, while the medial E cell generates two endoderm precursors, the lateral-most E cell is subject to an inductive interaction resulting in the generation of one endoderm and one mesoderm (the trunk lateral cell lineage) precursor (Shi and Levine, 2008). Later, during neural plate patterning, a muscle precursor is also generated from the lateral borders of the NN-lineage-derived neural plate (Nicol and Meinertzhagen, 1988; Nishida, 1987). Thus, as in other species, ascidian germ layer segregation can be viewed as a progressive process with part of the neural tissue arising from bipotential neuro-mesodermal progenitors (Henrique et al., 2015; Tzouanacou et al., 2009). The earliest cell divisions of the ascidian embryo along the A-V axis at the 8- and 32-cell stages can be considered as the earliest steps of germ layer segregation.
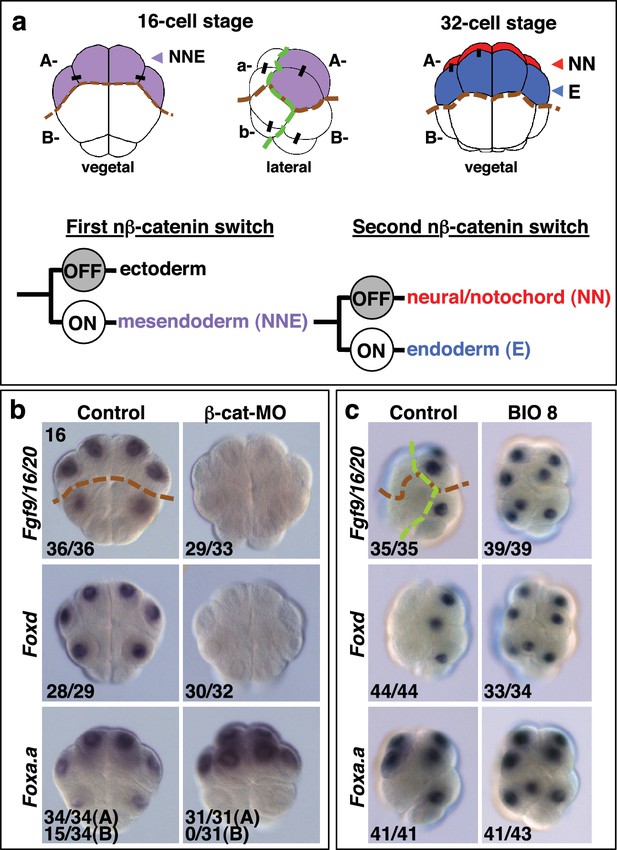
Foxa.a, Foxd and Fgf9/16/20 are candidate NNE lineage specification factors.
(a) Schematic drawings of embryos at the 16- and 32-cell stages. In this and all subsequent figures, where shown, a green dashed line separates the animal (ectoderm) from the vegetal (mesendoderm) hemispheres and a brown dashed line separates A- (A4.1) and a- (a4.2) lineages from B- (B4.1) and b- (b4.2) lineages. Different embryonic founder lineages are indicated on the drawings. NN and E cells are indicated in red and blue, respectively. Below the embryo drawings is a schematic representation of the two rounds of nβ-catenin-driven binary fate decisions that segregate firstly the mesendoderm lineages from the ectoderm lineages at the 16-cell stage and secondly segregate the mesoderm (NN) lineages from the endoderm (E) lineages at the 32-cell stage (Hudson et al., 2013). (b, c) Embryos analysed at the 16-cell stage for the marker indicated to the left of the panels following the treatment indicated above the panels [(b) vegetal pole view; (c) lateral view, vegetal pole to the right]. The numbers on the bottom-left corner of each panel indicate the proportion of embryos that the panel represents. The posterior most cells (at the bottom of the panels) are transcriptionally quiescent cells that will generate the germ line (Shirae-Kurabayashi et al., 2011). For Foxa.a expression in (b) control embryos showed expression in all four A-line (NNE) cells in 34/34 embryos, and in B-line cells, in 15/34 embryos, as indicated, whereas β-catenin-MO (β-cat-MO) injected embryos showed expression in NNE cells (31/31), but not B-line (0/31). Expression of Foxa.a in the four a-line precursors (not visible in the image) was not affected by β-catenin-MO injection.
β-catenin is a transcriptional co-activator which acts in a complex with TCF DNA-binding proteins to mediate the canonical Wnt signalling pathway (Valenta et al., 2012). The β-catenin/TCF complex promotes endoderm or mesendoderm in a wide range of organisms, and this process is therefore likely to represent an ancestral mechanism (Darras et al., 2011; Henry et al., 2008; Hudson et al., 2013; Imai et al., 2000; Logan et al., 1999; McCauley et al., 2015; Miyawaki et al., 2003; Momose and Houliston, 2007; Wikramanayake et al., 1998, 2003). We have previously shown that the earliest steps of germ layer segregation in ascidian embryos are mediated by two rounds of nuclear(n)-β−catenin-dependent binary fate decisions. The first nβ-catenin-driven binary fate decision takes place at the 8- to 16-cell stage. During this process, the β-catenin/TCF complex is differentially activated between mesendoderm and ectoderm progenitors, resulting in segregation of these lineages (Figure 1a) (Hudson et al., 2013; Oda-Ishii et al., 2016; Rothbächer et al., 2007). The second step takes place at the 32-cell stage and controls the segregation of NNE mesendoderm cells into endoderm (E cell) and notochord/neural (NN cell) lineages (Hudson et al., 2013). During this step, the β-catenin/TCF complex is again differentially activated between E and NN cells (Figure 1a). Therefore, cells in which nβ-catenin remains active during the two steps (ON + ON) are specified as endoderm lineage, cells in which nβ-catenin remains inactive during the two steps (OFF + OFF) are specified as ectoderm lineage and cells in which nβ-catenin is active during the first step but inactive during the second step (ON + OFF) are specified as notochord-neural lineage (Hudson et al., 2013). These two rounds of nβ-catenin-driven switches result in transcriptional activation of the lineage specifiers, Zic-related.b (Zic-r.b, formally ZicL) and Lhx3/4 (formally Lhx3), in NN and E cells, respectively (Imai et al., 2002c; Satou et al., 2001). One of the key features of these reiterative nβ-catenin-driven binary fate decisions is that the same asymmetric cue (nβ-catenin) is interpreted differently during each step (Bertrand and Hobert, 2010). Thus, in the NNE lineage, it is likely that the transient regulatory state induced by the first nβ-catenin input in NNE cells confers a distinct transcriptional response to the second nβ-catenin input on E cells.
In this study, we characterise the NNE lineage specification factors, which are induced by the first nβ-catenin input and address how these mesendoderm factors feed into the gene regulatory network of the NN and E lineages.
Results
Foxa.a, Foxd and Fgf9/16/20 are nβ-catenin transcriptional targets in NNE cells
Following the first nβ-catenin activation at the 16-cell stage, Foxa.a, Foxd, Fgf9/16/20, cadherinII and βCD1 (β-catenin downstream gene 1) are induced in the NNE cells, with at least Foxd and Fgf9/16/20 being direct targets of the β-catenin/Tcf7 complex (Imai, 2003; Imai et al., 2002a, 2002b, 2002c; Kumano et al., 2006; Oda-Ishii et al., 2016; Rothbächer et al., 2007; Satou et al., 2001). Consistent with a recent study (Oda-Ishii et al., 2016), we confirmed that in β-catenin–inhibited (β-catenin-MO injected) embryos analysed at the 16-cell stage, Foxd and Fgf/9/16/20 expression was lost (Figure 1b). In addition to the mesendoderm lineages, Foxa.a is also expressed in the a-line anterior ectoderm lineages in a nβ-catenin-independent fashion (Figure 1b,c) (Lamy et al., 2006). In β-catenin–inhibited embryos, Foxa.a expression persisted in NNE and a-lineage cells, probably due to transformation of vegetal cells into animal cells that has been reported previously (Figure 1b) (Imai et al., 2000; Oda-Ishii et al., 2016). Conversely, ectopic stabilisation of nβ-catenin resulted in activation of all three genes in ectoderm lineages at the 16-cell stage (Figure 1c). This was achieved by treating embryos with BIO, a chemical inhibitor of the upstream inhibitory regulator of β-catenin, GSK-3, from the eight-cell stage (Meijer et al., 2003). Thus, our results confirm that Foxd, Foxa.a and Fgf9/16/20 are transcriptional targets of nβ-catenin in vegetal cells, although Foxa.a also has a nβ-catenin-independent expression in a-line animal cells.
Foxa.a, Foxd and Fgf9/16/20-signals are required for the correct initiation of both NN and E gene expression
It is likely that these gene products, activated by the first nβ-catenin signal in NNE cells, act together with the second differential nβ-catenin signal to activate the distinct gene regulatory networks between NN and E cells. Consistent with this idea, Foxa.a has been shown to be required for both NN lineage and endoderm gene expression (Imai et al., 2006), with Foxd specifically required for NN lineage, but not endoderm fates, and Fgf9/16/20 contributing to notochord induction from the NN lineage (Imai et al., 2002a, 2002b; Yasuo and Hudson, 2007). However, we found that inhibiting any one of these factors prevented the correct initiation of gene expression in both NN (Zic-r.b) and E (Lhx3/4) lineages (Figure 2a, Table 1). We inhibited these factors using Morpholino anti-sense oligonucleotides (Foxd-MO, Foxa.a-MO, Fgf9-MO) and analysed Zic-r.b and Lhx3/4 expression at the 32-cell stage, when NN and E cell lineages become segregated. FGF signals are frequently mediated by the MEK/ERK signalling pathway, leading to transcriptional activation via ETS family transcription factors, as is the case in ascidian embryos (Bertrand et al., 2003; Kim and Nishida, 2001; Miya and Nishida, 2003; Yasuo and Hudson, 2007). We confirmed that Fgf9/16/20 is responsible for the broad activation of ERK at the 32-cell stage in most vegetal lineages, including NN and E lineages, as well as two neural lineages in the ectoderm (Figure 2—figure supplement 1f). Treatment of embryos from the 16-cell stage with the MEK inhibitor U0126, also inhibits this ERK1/2 activation (Kim and Nishida, 2001; Picco et al., 2007). Inhibition of Fgf9/16/20, MEK or ETS1/2 (ETS1/2-MO) gave similar results, although inhibition of ETS1/2 gave only a weak down-regulation of Zic-r.b expression at the 32-cell stage, perhaps indicating the involvement of additional transcription factors that are also known to mediate FGF signals in Ciona embryos (Figure 2a; Table 1) (Bertrand et al., 2003; Gainous et al., 2015). Maintenance of Foxa.a, Foxd and Fgf9/16/20 expression at the 32-cell stage is independent of each other (Figure 2—figure supplement 1a), as was shown previously for Foxd and Fgf9/16/20 in Ciona savigni embryos (Imai et al., 2002a).
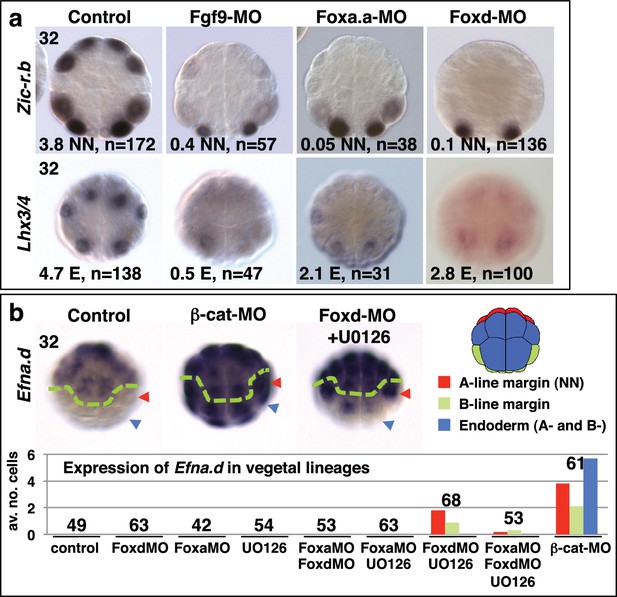
Foxa.a, Foxd and Fgf9/16/20 are required for initiation of NN and E gene expression.
(a) Embryos analysed at the 32-cell stage. The marker analysed is indicated on the left of the panels and the treatment indicated above the panels. The average number of NN (Zic-r.b) or E (Lhx3/4) cells expressing detectable levels of each gene is indicated. This remaining expression was generally weaker than control level expression. ‘n=’ represents the number of embryos analysed. (b) Expression of Efna.d under the conditions indicated. Embryos are shown in notochord-side view, animal pole up. The graph shows the average number of cells expressing Efna.d in different vegetal lineages at the 32-cell stage, as indicated by the key. All embryos showed ectoderm expression. The number of embryos analysed is indicated above the bars on the graph.
Expression of Zic-r.b in NN cells and Lhx3/4 in E cells of 32-cell stage embryos, following inhibition of Fgf-signalling components.
| Control | U0126 | ETS1/2-MO | |
|---|---|---|---|
| Zic-r.b NN cell | 4.0 cells (n = 163) | 0.75 cells (n = 58) | 3.4 cells* (n = 74) |
| Lhx3/4 E cell | 3.9 cells (n = 153) | 2.1** cell (n = 45) | 0.7 cells (n = 92) |
-
* 44/74 embryos exhibited weaker levels of Zic-r.b expression compared to controls.
-
**Remaining expression was weaker than control levels of expression.
In FGF-inhibited embryos, Zic-r.b expression recovered at the 64-cell stage (Figure 2—figure supplement 1a) (Imai et al., 2006; Kumano et al., 2006). Zic-r.b expression at the 32- and 64-cell stages can be mediated by separate enhancer elements (Anno et al., 2006). In addition, in the NN-cell lineage, FGF-signalling is required for notochord fate, but has to be attenuated for neural fate (Minokawa et al, 2001; Picco et al., 2007; Yasuo and Hudson, 2007). Thus, an FGF-independent expression of Zic-r.b at the 64-cell stage, at least in neural fated cells, is not unexpected. In Foxa.a- and Foxd- inhibited embryos, Zic-r.b continues to be repressed in the NN-cell lineages at the 64-cell stage (Figure 2—figure supplement 1a) and later (Imai et al., 2006), consistent with a requirement for Foxa.a and Foxd for both NN cell lineage-derived structures, the notochord and caudal central nervous system (CNS) (Imai et al., 2002b, 2006).
Endoderm gene expression was continuously reduced up to at least the early gastrula stage, following inhibition of any one of the NNE factors (Figure 2—figure supplement 1a–e). However, using alkaline phosphatase activity as an indicator of endoderm formation (Whittaker, 1977), a complete loss of endoderm at larval stages was observed only in Foxa.a-inhibited embryos, consistent with previous studies (Figure 2—figure supplement 2) (Imai et al., 2002b, 2006). In Foxd and FGF-signal inhibited embryos, a large domain of alkaline phosphatase activity could be detected, suggesting that endoderm fate recovers in these embryos (Figure 2—figure supplement 2). Simultaneous repression of Foxd and FGF-signalling, however, resulted in both a stronger repression of early endoderm gene expression as well as an almost complete absence of alkaline phosphatase activity at larval stages (Figure 2—figure supplement 1–2). Thus, for eventual endoderm formation, the embryo is able to compensate for loss of either Foxd or FGF-signals but is not able to compensate for loss of both.
As well as promoting vegetal ‘mesendoderm’ fates, nβ-catenin also represses the ectoderm gene programme in vegetal cells (Hudson et al., 2013; Imai et al., 2000; Oda-Ishii et al., 2016; Rothbächer et al., 2007). In β-catenin knock-down embryos, ectopic expression of the early ectoderm gene Efna.d (formally ephrin-Ad) is observed in both NN and E cells at the 32-cell stage (Figure 2b) (Hudson et al., 2013) as well as in NNE cells at the 16-cell stage (Oda-Ishii et al., 2016). Double inhibition of Foxd and FGF-signals also resulted in ectopic expression of Efna.d in NN cells, but never in E cells (Figure 2b). Thus, NNE factors repress the ectoderm genetic programme in NN cells. A lack of derepression in E cells is probably due to the presence of nβ-catenin in E cells at the 32-cell stage (Hudson et al., 2013), suggesting that nβ-catenin can repress the ectoderm genetic programme both via and independently of the NNE factors.
Combinatorial activity of Foxa.a, Foxd and Fgf9/16/20 induces a mesendoderm state
Our data so far show that Foxa.a, Foxd and Fgf9/16/20-ERK1/2 are individually required for the correct initiation of the genetic programmes of both NN and E cell lineages. Indeed, co-expression of these three factors takes place only in mesendoderm lineages, the NNE and B-line mesendoderm lineages of the 16- and 32-cell stage embryo (Figure 1; Figure 2—figure supplement 1a). At the 32-cell stage, the E cells continue to express these three genes, while NN cells express only Foxa.a and Fgf9/16/20. However, we have previously shown that Foxd transcripts preferentially segregate into the NN cells during the NN-E cell division, before they rapidly disappear (Hudson et al., 2013). Thus, NN cells also contain Foxd transcripts early in their cell cycle. We conclude that Foxa.a, Foxd and Fgf9/16/20 are co-expressed only in mesendoderm lineages. We next addressed whether these three factors were sufficient to induce a mesendoderm regulatory state.
As well as in vegetal cells, Foxa.a is also expressed in a-line anterior animal cells and ERK1/2 is activated in one pair of a-line cells (the a6.5 pair) at the 32-cell stage (Figure 3a) (Hudson et al., 2003; Lamy et al., 2006). Thus, a6.5 cells possess two of the three mesendoderm lineage specifiers and yet they do not adopt an NNE-like lineage. Consistent with the notion that coexpression of Foxa.a, Foxd and Fgf9/16/20 represents a NNE regulatory state, reintroduction of the remaining factor, Foxd, by mRNA injection, was able to convert a-line cells to a mesendoderm state (Figure 3). Injection of Foxd mRNA resulted in ectopic expression of Zic-r.b in a-line cells at the 32-cell stage (Figure 3b) (Imai et al., 2002c). Similarly, expression of Bra, a marker of notochord precursors, was induced at the 64-cell stage (Figure 3b). The broad ectopic expression of Zic-r.b and Bra in the a-lineage was clearly not restricted to the a6.5 cells. The most likely reason for this was that Foxd mRNA injection led to weak activation of Fgf9/16/20 and strong inhibition of Efna.d expression in the ectoderm cells (Figure 3—figure supplement 1). Efna.d is a known antagonist of FGF-signals in Ciona and its inhibition results in widespread activation of ERK1/2 in ectoderm lineages (Ohta and Satou, 2013; Picco et al., 2007; Shi and Levine, 2008). Consistent with this, when Foxd mRNA-injected embryos were treated with U0126, the ectopic expression of Zic-r.b was reduced and Bra expression was completely suppressed, mimicking the effect of MEK inhibition on endogenous Zic-r.b and Bra gene expression (Figure 3b). Thus, injection of Foxd mRNA is sufficient to convert Foxa.a/ERK1/2-positive a-line cells into mesoderm.
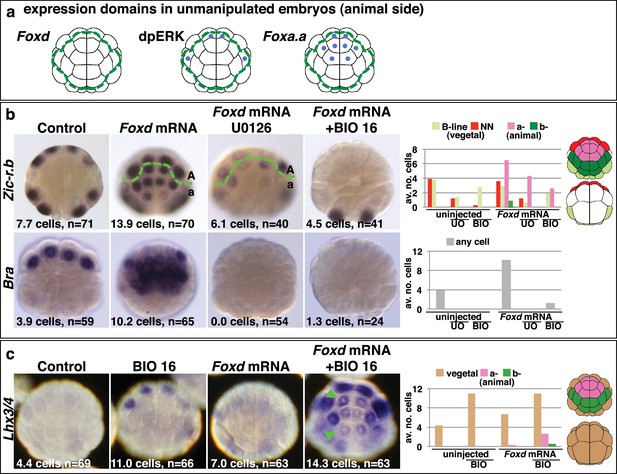
Creation of an ectopic Foxa.a, Foxd, FGF-signal overlap leads to ectopic mesendoderm formation.
(a) Schematics show endogenous ectodermal expression of Foxa.a, Foxd (no expression) and activation of ERK (dpERK), indicated by blue dots. (b–c) Treatment is indicated above the panels and marker analysed to the left. All embryos in animal pole view except control Bra (vegetal pole view). Numbers show the total average number of cells per embryo expressing each marker. n = total number of embryos analysed. The graphs show the average number of cells expressing each maker in the lineages indicated on the keys, following the treatments indicated of the x-axis. No Zic-r.b expression was detected in endoderm lineages. In (c), the green arrowheads highlight the eight a-lineage cells. For (b), representative panels of uninjected/UO-treated and uninjected/BIO-treated embryos are not shown. The numbers of these experiments are: for Zic-r.b- U0126 alone n = 40 (average number of cells 2.6), BIO-16 alone n = 40 (average number of cells 2.9) and for Bra- U0126 alone, n = 39 (average number of cells 0.0); BIO-16 alone n = 31 (average number of cells 0.0).
Injection of Foxd mRNA, however, was not sufficient to induce endoderm gene expression in ectoderm cells (Figure 3c). This was expected since the two-step nβ-catenin binary fate decision model predicts that segregation of the endoderm lineage from the NNE lineage requires a second round of nβ-catenin activation (Figure 1a) (Hudson et al., 2013). Accordingly, when Foxd-mRNA injected embryos were treated from the late 16-cell stage with BIO in order to mimic the second input of nβ-catenin activation, both Zic-r.b and Bra were repressed and Lhx3/4 ectopically activated in the a-line ectoderm cells (Figure 3b–c). Thus, the NNE-like state induced in animal cells by Foxd mRNA injection behaves in the same way as NNE state of unmanipulated embryos.
Taken together, these experiments provide strong evidence that the combinatorial activity of Foxa.a, Foxd and Fgf9/16/20-ERK1/2 represents a NNE mesendoderm regulatory state downstream of the first round of nβ-catenin input. To further test this model, we addressed whether co-expression of Foxa.a, Foxd and Fgf9/16/20 was sufficient to rescue mesoderm in β-catenin-knockdown embryos. β-catenin-MO injected embryos would express only Foxa.a among the three genes (Figure 1b). We have shown that injection of Foxd mRNA results in induction of low levels of Fgf9/16/20 expression, together with a strong suppression of Efna.d expression (Figure 3—figure supplement 1). Thus, injection of Foxd mRNA should be sufficient to recapitulate a Foxd/Foxa.a/Fgf9/16/20 overlap. Consistent with this, injection of Foxd mRNA was able to rescue expression of NN-lineage genes (Zic-r.b and Bra) in β-catenin-MO embryos and, as expected, this recovery depended on an intact FGF-signalling pathway (Figure 4). We conclude that co-expression of Foxd+Foxa.a+Fgf9/16/20 is sufficient to induce a mesendoderm regulatory state, which can then be further programmed into mesoderm or endoderm lineage by manipulation of nβ-catenin activity.
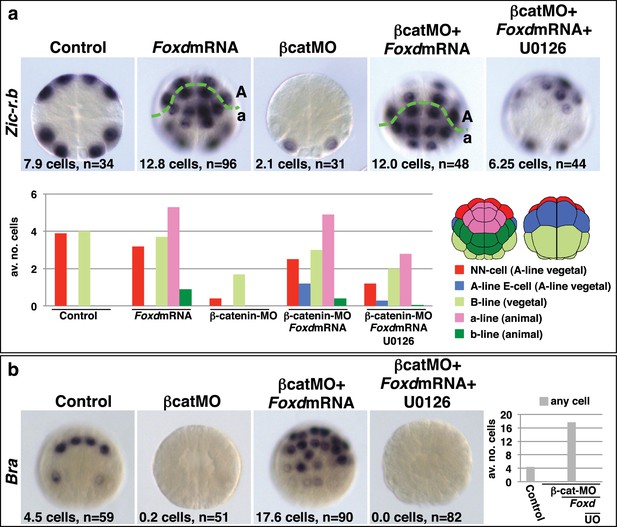
Foxd mRNA injection rescues mesoderm in β-catenin-MO injected embryos.
(a–b) Treatment is indicated above the panels and marker analysed to the left of the panels. The total average number of cells per embryo is indicated, ‘n=’ indicates the total number of embryos analysed for each treatment. The graphs show the average number of cells expressing each marker in the lineages indicated by the keys, following the treatments indicated.
Foxa.a, Foxd and Fgf9/16/20 act synergistically to reprogramme developing ectoderm cells to a mesendoderm state
We next addressed whether ectopic expression of Foxd, Foxa.a and Fgf9/16/20 was able to reprogramme developing ectoderm to a mesendoderm state (Figure 5, Figure 5—figure supplement 1). The upstream regulatory sequences of the Fucosyltransferase-like (FT) gene become active in ectoderm cells from the 64-cell stage, when the ectoderm genetic programme is already underway (Figure 5—figure supplement 1) and when these cells no longer express Foxa.a, Foxd or Fgf9/16/20 (Imai et al., 2004; Pasini et al., 2012). Using FT promoter-driven constructs (pFT>Foxa.a, pFT>Foxd and pFT>Fgf9/16/20), we expressed Foxa.a, Foxd and Fgf9/16/20 in different combinations in ectoderm lineages. To simplify the analysis and to rule out the possibility that signals from the vegetal cells may influence the experimental outcome, animal hemispheres of electroporated embryos were isolated by micro-dissection at the eight-cell stage. Isolated explants were cultured until the neurula stage when they were assayed for Bra expression (Figure 5a–b). Bra was chosen for this assay for its mesoderm (notochord)-specific expression. We observed a clear combinatorial effect between Foxa.a, Foxd and Fgf9/16/20 on the reprogramming of ectoderm to mesoderm, with strong induction of Bra seen only when all three constructs were co-electroporated (Figure 5b). This reprogramming was accompanied by a strong downregulation of ectoderm gene expression and ectopic expression of Zic-r.b in the ectoderm cells of whole embryos (Figure 5—figure supplement 1). Furthermore, ectoderm explants could be reprogrammed to adopt an endoderm state (Figure 5c). To achieve this, ectoderm explants from embryos electroporated with the triple combination (pFT>Foxa.a, pFT>Foxd and pFT>Fgf9/16/20) were treated with a pulse of BIO from the 76-cell stage to mimic the second round of nβ-catenin activation that normally drives the segregation of NN and E lineages (Figure 5c). The 76-cell stage was chosen, as at this stage ectopic expression driven by the pFT constructs is readily detectable (Figure 5—figure supplement 1a), the ectoderm programme is downregulated (Figure 5—figure supplement 1b) and ectopic Zic-r.b is not yet detected (Figure 5—figure supplement 1c). This stage thus represented the best approximation of the NNE state of normal embryos. We confirmed that BIO-treatment was able to induce nuclear translocation of β-catenin in isolated explants at the 76-cell stage (Figure 5—figure supplement 2). Endoderm induction was assayed by detection of Lhx3/4 expression at the mid-gastrula stage, that is approximately 1 hr after the onset of BIO-treatment. Coupling these three factors with BIO-treatment resulted in strong induction of Lhx3/4 expression. We conclude that the combinatorial activity of Foxa.a, Foxd and Fgf9/16/20 is sufficient to reprogramme developing ectoderm cells to adopt a mesendoderm state.
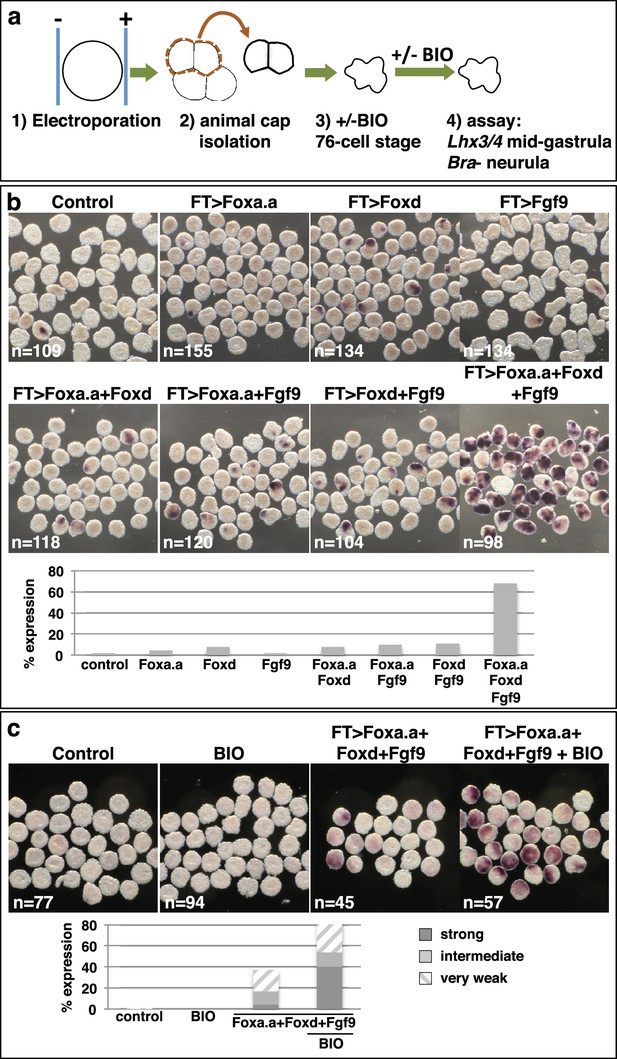
Reprogramming the ectoderm lineage to mesendoderm.
(a) Experimental scheme. Embryos were electroporated and the ectoderm lineage (animal cap) isolated at the eight-cell stage. Ectodermal explants were cultured until the mid-gastrula stage for Lhx3/4 expression or until the neurula stage for Bra expression. Optionally, explants were treated with BIO, when control sibling embryos reached the 76-cell stage, for approximately 1 hr prior to fixation (Lhx3/4 only). (b–c) Expression of Bra (b) and Lhx3/4 (c) in isolated ectodermal explants, following the treatments indicated above the panels. ‘n=’ represents the number of explants analysed. Graphs shows the percentage of explants with any level of Bra expression or level of Lhx3/4 expression indicated by the key, under various conditions (Foxa.a= pFT>Foxa.a; Foxd = pFT>Foxd; Fgf9= pFT>Fgf9/16/20; control = unelectroporated).
Discussion
In this study, we have identified Foxa.a, Foxd and Fgf9/16/20 as the mesendoderm lineage specifiers of the NNE cell. Transcriptional activation of Foxa.a, Foxd and Fgf9/16/20 is induced by the first nβ-catenin switch (Figures 1a,6a). Co-expression of these three factors is sufficient to reprogramme ectoderm cells to adopt a mesendoderm state. This ectopic mesendoderm state can be further converted into either mesoderm or endoderm by modulating nβ-catenin activation.
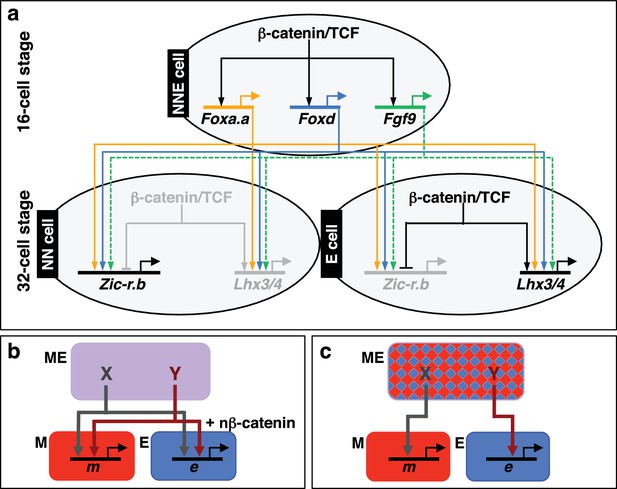
Gene regulatory model for segregation of NNE into NN and E lineages.
(a) Each factor induced by nβ-catenin activation at the 16-cell stage feeds into both the NN and E lineage genes. The dashed line for Fgf9/16/20 represents a signalling molecule (most likely mediated, at least in part, by Ets1/2 transcription factor (Table 1). Differential gene expression between NN and E cells is mediated by the second nβ-catenin-driven switch. (b–c) Schematic regulatory architectures during mesendoderm segregation. ME = mesendoderm lineage; M = mesoderm lineage; E = endoderm lineage; e = endoderm gene; m = mesoderm gene; X, Y = genes expressed in mesendoderm cells. (b) Ascidian and nematode mesendoderm regulatory architecture. (c) ‘Mixed-lineage’ mesendoderm regulatory architecture.
A model for ascidian germ layer segregation
We propose the following model to summarise the initial stages of germ layer segregation in ascidian embryos (Figure 6a). At the 8- to 16-cell stage of development, nβ-catenin, activated specifically in vegetal cells by as yet unknown mechanisms, promotes Foxa.a, Foxd and Fgf9/16/20 expression and represses ectoderm gene expression (Hudson et al., 2013; Imai et al., 2000; Oda-Ishii et al., 2016; Rothbächer et al., 2007). Foxa.a, Foxd and Fgf9/16/20, are co-expressed exclusively in mesendoderm lineages at the 16- to 32-cell stage of development (Imai et al., 2002a, 2002b; Oda-Ishii et al., 2016), where they are required, individually, for the correct initiation of both NN and E cell lineage gene expressions at the 32-cell stage (Figure 2a). The NNE factors are also required to repress ectoderm gene expression: co-inhibition of Foxd and Fgf9/16/20 resulted in ectopic ectoderm gene expression in NN cells (Figure 2b) and Foxd overexpression alone was able to repress ectoderm gene expression (Figure 3—figure supplement 1). However, our data also suggest that nβ-catenin can repress ectoderm gene expression independently of these three factors (Figure 2b). Recently, it has been shown that this can take place via a physical interaction between β-catenin/Tcf7 and Gata.a, preventing this key regulator of ectoderm lineage from binding to its DNA target sites (Oda-Ishii et al., 2016; Rothbächer et al., 2007).
Following inhibition of Foxa.a, Foxd or Fgf9/16/20, both endoderm and mesoderm development is perturbed at later stages of development, although there is a redundancy between Foxd and FGF-signalling for the eventual recovery of endoderm (Figure 2 and Figure 2—figure supplement 1, Figure 2—figure supplement 2) (Imai et al., 2002a, 2002b, 2002c, 2006; Kumano et al., 2006). It is likely that these factors play on-going roles during mesoderm and endoderm lineage progression. For example, ERK1/2 activity is detected in both notochord and endoderm until the early gastrula stage (Nishida, 2003; Yasuo and Hudson, 2007), Fgf9/16/20 is required at the 64-cell stage for induction of notochord and repression of neural gene expression in the notochord lineage (Imai et al., 2002a; Kim and Nishida, 2001; Minokawa et al., 2001; Yasuo and Hudson, 2007) and Foxa.a is continuously expressed in notochord and endoderm, suggesting an on-going role for Foxa.a in both of these lineages (Imai et al., 2004).
Importantly, creating ectopic zones of co-expression of these three factors in distinct embryological settings, revealed their strong synergistic ability to induce a mesendoderm state, which can be further programmed to an NN or E-like lineage by modulation of nβ-catenin levels (Figures 3–5 and Figure 3—figure supplement 1, Figure 5—figure supplement 1, Figure 5—figure supplement 2). We conclude that Foxa.a, Foxd and Fgf9/16/20 are crucial for the mesendoderm ground state that canalises the daughter lineages to adopt either E or NN fates depending on the status of the second nβ-catenin input.
It is important to bear in mind that the germ layers are still not fully segregated at the 32-cell stage. While this manuscript has focused on the mesendoderm fates that arise from the NNE lineage, this lineage also produces neural tissue. NNE cells divide into E cells and NN cells. In addition to notochord, the NN cell generates the posterior part of the CNS, including the equivalent of the ‘spinal cord’ of vertebrates (reviewed in [Hudson, 2016]). The binary cell fate decision between neural and notochord takes place at the 64-cell stage (Minokawa et al., 2001; Picco et al., 2007). The lateral neural progenitors that arise from the NN-cell lineage also produce a muscle cell during neural plate patterning, following another neuromesodermal binary fate decision (reviewed in [Hudson and Yasuo, 2008]). Bipotential neuromesoderm progenitors are not an ascidian novelty (Henrique et al., 2015; Tzouanacou et al., 2009). For example, in the zebrafish tailbud, bipotential neuromesodermal progenitor cells generate notochord and floorplate (ventral spinal cord) (Row et al., 2016) and in both human and mouse embryonic stem cells and zebrafish tailbud stem cells, bipotential neuromesodermal progenitors generate paraxial mesoderm and posterior neural tube (Gouti et al., 2014; Martin and Kimelman, 2012). Even in the classical mesendoderm model, that is the C. elegans EMS cell, the MS (mesoderm) lineage also gives rise to some neurons (Sulston and Horvitz, 1977; Sulston et al., 1983). The lateral E cells of Ciona are also not yet fate-restricted to endoderm fate. At the 64-cell stage of development, the lateral E cell divides into one endoderm and one trunk lateral cell (mesenchyme) precursor, following induction of trunk lateral cell fate (Shi and Levine, 2008). Thus, as in other species, ascidian germ layer segregation is an progressive process (Tzouanacou et al., 2009) and NNE specification should thus be considered as its first step.
Regulatory architectures of mesendoderm
We have shown that, in ascidian embryos, individual mesendoderm lineage specifiers are required for the initiation of both mesoderm and endoderm GRNs (Figure 6). Furthermore, we have shown that the combinatorial activity of just three NNE factors is sufficient to reprogramme developing ectoderm cells to a mesendoderm state. The mesendoderm regulatory state in ascidian embryos is similar to the situation in the C. elegans EMS cell in which the MED1/2 GATA factors feed into both E (endoderm) and MS (mesoderm) lineage specification, such that MED1/2 directly activates both MS and E target genes (Broitman-Maduro et al., 2005; Maduro et al., 2001, 2015; McGhee, 2013). Similarly, Foxa.a and Foxd can bind to the upstream sequences of both Zic-r.b (NN lineage) and Lhx3/4 (E lineage), suggesting that this genetic interaction is direct (Kubo et al., 2010). While there is little doubt that mesendoderm transiently forms during embryogenesis of many animal models, and that both mesoderm and endoderm are induced by similar upstream regulators (β-catenin in invertebrates, β-catenin and Nodal in vertebrates), in most cases the transcriptional nature of the mesendoderm state does not appear to be similar to that of ascidians or nematodes. In particular, the existence of mesendoderm lineage specifiers (that is individual factors required for the initiation of both mesoderm and endoderm GRNs) have not been described in the majority of model organisms. For example, in sea urchins and anamniote vertebrates, mesendoderm has been described as a mixed regulatory state with simultaneous activation of mesoderm and endoderm GRNs, prior to the lineage segregation of these fates (Peter and Davidson, 2010; Rodaway and Patient, 2001). This type of ‘mixed-lineage’ regulatory architecture is also described in other systems and displays characteristics of multi-lineage priming, whereby the GRNs of two lineages are simultanously activated prior to lineage segregation (Figure 6c) (Graf and Enver, 2009; Nimmo et al., 2015). If this were the scenario for the ascidian mesendoderm regulatory state, one would expect individual NNE factors to be required for, and be able to induce, only one or other of the two subsequent lineages (NN or E), but not both. A ‘mixed-lineage’ regulatory architecture is therefore not consistent with our data describing the NNE mesendoderm regulatory state (Figure 6).
These two regulatory architectures are, however, unlikely to be mutually exclusive. In sea urchin and sea stars, for example, genes interacting with both mesoderm and endoderm GRNs have been identified (http://sugp.caltech.edu/endomes/) (Davidson et al., 2002; McCauley et al., 2015). It cannot be ruled out that mesendoderm lineage specifiers, acting upstream of both endoderm and mesoderm GRNs, are more broadly utilised, but are simply difficult to uncover due to the sheer complexity of early embryos and their GRNs (Ben-Tabou de-Leon and Davidson, 2009; Kiecker et al., 2016; Tremblay, 2010). It is also possible that the regulatory architecture of nematode and ascidian mesendoderm resulted from an adaption to a lineage-based mode of development with small numbers of cells, perhaps enabling these rapidly developing embryos to bypass the need for cross-repression and prolonged stabilisations of the endoderm and mesoderm GRNs. In summary, it is not yet clear whether an obligate mesendoderm state (that is a state with mesendoderm lineage specifiers) is present in the majority of bilaterian developmental programmes, although this seems to be the case in nematode and ascidian embryos.
Materials and methods
Overexpression and knockdown tools
Request a detailed protocolMorpholinos (MOs) were purchased from GeneTools (Philomath, Oregon) and have been reported previously: β-catenin-MO (Hudson et al., 2013); Foxa.a-MO and Foxd-MO (Imai et al., 2006); Fgf9/16/20-MO and Fgf8/17/18-MO (Yasuo and Hudson, 2007), ETS1/2-MO (Bertrand et al., 2003). Foxa.a-MO was injected at 0.85 mM and ETS1/2-MO at 0.75 mM. All other morpholinos were injected at 0.5 mM. U0126 was used at 2 μM and BIO (GSK-3 inhibitor IX) at 2.5 μM (both were purchased from Calbiochem (Merck, Darmstadt, Germany)). Since a full-length cDNA clone for Ciona intestinalis Foxd is not available in gene collection plates (Gilchrist et al., 2015; Satou et al., 2002), we synthesised the Ciona savignyi Foxd mRNA from pRN3-Cs-Foxd (Imai et al., 2002b) using mMESSEGEmMACHINE kit (Thermo Fisher Scientific, Waltham, MA). The Ciona savigni Foxd used in this study corresponds to Genbank accession number AB057738.1. Foxd mRNA was injected at 75ng/μl. In order to generate pFT>Foxa.a, pFT>Foxd and pFT>Fgf9/16/20, we first constructed Gateway (Invitrogen, a brand of Thermo Fisher Scientific) pENTR clones containing ORFs of these genes. ORFs of Cs-Foxd, Foxa.a and Fgf9/16/20 were PCR-amplified using the following primer pairs and templates: Foxa.a-attB1 (aaaaagcaggctaccATGATGTTGTCGTCTCCACC) and Foxa.a-attB2 agaaagctgggtTTAGCTTGCTGGTACGCAC) on cicl044j20 template; FGF9-attB1 (aaaaagcaggctaccATGTCTATGTTAACCAACATGTTAGG) and FGF9-attB2 (agaaagctgggtTCAGTAGAGTCGGCCAGAGTAC) on citb007k01; CsFoxd-attB1 (aaaaagcaggctaccATGACTGTGGACTCTTGTACAG) and CsFoxd-attB2 (agaaagctgggtCTAAATAAGTTTATACGGGAATGG) on pRN3-Cs-Foxd. The Fucosyltransferase-like driver has been reported previously (Pasini et al., 2012). The promoter region was PCR-amplified using the following pair of primers to generate a destination vector pSP1.72BSSPE-pFT::RfA-venus (Roure et al., 2007): pFT-attB3 (ggggacaagtttgtataataaagtaggctGGCATCATAACGTACAACCTG) and pFT-attB5 (ggggaccactttgtatacaaaagttgggtTGCAGCGGTAGAGTTTACTATTATC). pFT>Foxa.a, pFT>Foxd and pFT>Fgf9/16/20 were then generated by LR reaction between corresponding pENTR clones and pSP1.72BSSPE-pFT::RfA-venus.
Embryological experiments
Request a detailed protocolAdult Ciona intestinalis were purchased from the Station Biologique de Roscoff (France). Blastomere names, lineage and the fate maps are previously described (Conklin, 1905; Nishida, 1987). Ascidian embryo culture and microinjection have been described (Sardet et al., 2011). All microinjections were carried out in unfertilised eggs. The electroporation protocol was based on Christiaen et al., 2009. Up to 60 μg of circular plasmid DNA was made up to 250 μl at 0.6 M mannitol. DNA/mannitol solution was mixed with 100 μl of eggs in artificial sea water supplemented with 0.5% BSA (to help prevent sticking). Electroporation was carried out at 50V for 16 ms using a BTX (Harvard Apparatus, Holliston, Massachusetts) ECM 830 and electroporated embryos transferred to agarose-coated dishes. For data shown in Figure 5—figure supplement 1a, 50 μg of pFT>Foxd was used. Otherwise, each FT construct was used at 20 μg to give a maximum total of 60 μg. pFT>tdTomato was used as a control electroporation at 60 μg. In all experiments, embryos that failed to develop were discarded and all other embryos scored. All data were pooled from at least two independent experiments (i.e., on different batches of embryos).
The experimental design of the BIO treatment of electroporated ectodermal explants, shown in Figure 5c, is as follows. Embryos were electroporated with three plasmids, pFT>Foxa.a + pFT>Foxd + pFT>Fgf9/16/20. Ectoderm explants were isolated at the eight-cell stage from control (unelectroporated) and electroporated embryos. Each sample of explants was split into two groups and one group of each treated with BIO from the 76-cell stage. BIO treatment was continued until fixation, when sibling embryos reached the mid-gastrula stage. Explants remained in BIO for approximately 1 hr at 20°C.
In situ hybridisation, gene naming, alkaline phosphatase staining and dpERK and β-catenin immunofluorescence
Request a detailed protocolAll gene markers used for in situ hybridisation have previously been described (Hudson et al., 2013; Imai et al., 2004) (http://ghost.zool.kyoto-u.ac.jp). According to recent nomenclature guidelines, we used Zic-r.b (previously called ZicL) to describe the five copies of ZicL gene named Zic-r.b – Zic-r.f, Lhx3/4 (previously Lhx3) and Efna.d (previously ephrin-Ad) (Stolfi et al., 2015). The in situ hybridisation and alkaline phosphatase staining protocols are previously described (Hudson et al., 2013). All single embryo panels, except those in Figure 3c, were mounted in 50–80% glycerol and photographed on an Olympus (Tokyo, Japan) BX51 using a Leica DFC310FX camera. All multi-embryo panels as well as single-embryo panels in Figure 3c were taken of embryos in PBT on a Leica (Leica microsystems, Vienna, Austria) Macroscope Z16 APO with a Canon (Tokyo, Japan) EOS 60D camera. The dpERK and β-catenin immunofluorescence protocols are described previously (Haupaix et al., 2014; Hudson et al., 2013). Immunostained embryos were mounted in Vectashield-DAPI (Vector laboratories, Burlingame, CA), analysed on a Leica SP5 confocal microscope and processed with Image J.
References
-
Transcriptional regulation of ZicL in the Ciona intestinalis embryoDevelopment Genes and Evolution 216:597–605.https://doi.org/10.1007/s00427-006-0080-9
-
Experimentally based sea urchin gene regulatory network and the causal explanation of developmental phenomenologyWiley Interdisciplinary Reviews: Systems Biology and Medicine 1:237–246.https://doi.org/10.1002/wsbm.24
-
Lineage programming: navigating through transient regulatory states via binary decisionsCurrent Opinion in Genetics & Development 20:362–368.https://doi.org/10.1016/j.gde.2010.04.010
-
Electroporation of Transgenic DNAs in the Sea Squirt CionaCold Spring Harbor Protocols 2009:pdb.prot5345.https://doi.org/10.1101/pdb.prot5345
-
BookThe Organisation and Cell Lineage of the Ascidian Egg, 13Philadelphia: Academy of Natural Sciences.https://doi.org/10.5962/bhl.title.4801
-
A genomic regulatory network for developmentScience 295:1669–1678.https://doi.org/10.1126/science.1069883
-
Diverse ETS transcription factors mediate FGF signaling in the Ciona anterior neural plateDevelopmental Biology 399:218–225.https://doi.org/10.1016/j.ydbio.2014.12.032
-
Neuromesodermal progenitors and the making of the spinal cordDevelopment 142:2864–2875.https://doi.org/10.1242/dev.119768
-
Similarity and diversity in mechanisms of muscle fate induction between ascidian speciesBiology of the Cell 100:265–277.https://doi.org/10.1042/BC20070144
-
The central nervous system of ascidian larvaeWiley Interdisciplinary Reviews: Developmental Biology.https://doi.org/10.1002/wdev.239
-
(beta)-catenin mediates the specification of endoderm cells in ascidian embryosDevelopment 127:3009–3020.
-
Regulatory blueprint for a chordate embryoScience 312:1183–1187.https://doi.org/10.1126/science.1123404
-
Early embryonic expression of FGF4/6/9 gene and its role in the induction of mesenchyme and notochord in Ciona savignyi embryosDevelopment 129:1729–1738.
-
An essential role of a FoxD gene in notochord induction in ciona embryosDevelopment 129:3441–3453.
-
Multiple functions of a Zic-like gene in the differentiation of notochord, central nervous system and muscle in Ciona savignyi embryosDevelopment 129:2723–2732.
-
Molecular specification of germ layers in vertebrate embryosCellular and Molecular Life Sciences 73:923–947.https://doi.org/10.1007/s00018-015-2092-y
-
Role of the FGF and MEK signaling pathway in the ascidian embryoDevelopment, Growth and Differentiation 43:521–533.https://doi.org/10.1046/j.1440-169X.2001.00594.x
-
Vertebrate mesendoderm induction and patterningCurrent Opinion in Genetics & Development 10:350–356.https://doi.org/10.1016/S0959-437X(00)00095-2
-
Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryoDevelopment 126:345–357.
-
MED GATA factors promote robust development of the C. elegans endodermDevelopmental Biology 404:66–79.https://doi.org/10.1016/j.ydbio.2015.04.025
-
The Caenorhabditis elegans intestineWiley Interdisciplinary Reviews: Developmental Biology 2:347–367.https://doi.org/10.1002/wdev.93
-
GSK-3-selective inhibitors derived from Tyrian purple indirubinsChemistry & Biology 10:1255–1266.https://doi.org/10.1016/j.chembiol.2003.11.010
-
Binary specification of nerve cord and notochord cell fates in ascidian embryosDevelopment 128:2007–2017.
-
Nuclear localization of beta-catenin in vegetal pole cells during early embryogenesis of the starfish Asterina pectiniferaDevelopment, Growth and Differentiation 45:121–128.https://doi.org/10.1034/j.1600-0854.2004.00681.x
-
Primed and ready: understanding lineage commitment through single cell analysisTrends in Cell Biology 25:459–467.https://doi.org/10.1016/j.tcb.2015.04.004
-
Spatio-temporal pattern of MAP kinase activation in embryos of the ascidian halocynthia roretziDevelopment, Growth and Differentiation 45:27–37.https://doi.org/10.1046/j.1440-169X.2003.00672.x
-
The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stageDevelopmental Biology 340:188–199.https://doi.org/10.1016/j.ydbio.2009.10.037
-
Embryological methods in ascidians: the villefranche-sur-mer protocolsMethods in Molecular Biology 770:365–400.https://doi.org/10.1007/978-1-61779-210-6_14
-
Early embryonic expression of a LIM-homeobox gene Cs-lhx3 is downstream of beta-catenin and responsible for the endoderm differentiation in Ciona savignyi embryosDevelopment 128:3559–3570.
-
Post-embryonic cell lineages of the nematode, caenorhabditis elegansDevelopmental Biology 56:110–156.https://doi.org/10.1016/0012-1606(77)90158-0
-
The embryonic cell lineage of the nematode Caenorhabditis elegansDevelopmental Biology 100:64–119.https://doi.org/10.1016/0012-1606(83)90201-4
-
Formation of the murine endoderm: lessons from the mouse, frog, fish, and chickProgress in Molecular Biology and Translational Science 96:1–34.https://doi.org/10.1016/B978-0-12-381280-3.00001-4
-
The many faces and functions of β-cateninEMBO Journal 31:2714–2736.https://doi.org/10.1038/emboj.2012.150
-
Segregation during cleavage of a factor determining endodermal alkaline phosphatase development in ascidian embryosJournal of Experimental Zoology 202:139–153.https://doi.org/10.1002/jez.1402020202
-
beta-Catenin is essential for patterning the maternally specified animal-vegetal axis in the sea urchin embryoProceedings of the National Academy of Sciences of the United States of America 95:9343–9348.https://doi.org/10.1073/pnas.95.16.9343
Article and author information
Author details
Funding
Centre National de la Recherche Scientifique
- Hitoyoshi Yasuo
Université Pierre et Marie Curie
- Hitoyoshi Yasuo
Fondation ARC pour la Recherche sur le Cancer (#1144)
- Hitoyoshi Yasuo
Agence Nationale de la Recherche (ANR-09-BLAN-0013-01)
- Hitoyoshi Yasuo
Fondation ARC pour la Recherche sur le Cancer (PJA 20131200223)
- Hitoyoshi Yasuo
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank U Röthbacher and P Lemaire (FT promoter), H Nishida (β-catenin antibody), Y Satou (pRN3-Cs-Foxd and Foxa.a-MO) and N Satoh (Gene Collection Plates) for tools, Evelyn Houliston and David McClay for critical reading of the manuscript and helpful discussions and Jenifer Croce and David McClay for interesting discussions on sea urchin mesendoderm GRNs.
Version history
- Received: January 25, 2016
- Accepted: June 24, 2016
- Accepted Manuscript published: June 28, 2016 (version 1)
- Accepted Manuscript updated: June 29, 2016 (version 2)
- Accepted Manuscript updated: June 30, 2016 (version 3)
- Version of Record published: July 14, 2016 (version 4)
Copyright
© 2016, Hudson et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,136
- views
-
- 262
- downloads
-
- 37
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Computational and Systems Biology
- Developmental Biology
Organisms utilize gene regulatory networks (GRN) to make fate decisions, but the regulatory mechanisms of transcription factors (TF) in GRNs are exceedingly intricate. A longstanding question in this field is how these tangled interactions synergistically contribute to decision-making procedures. To comprehensively understand the role of regulatory logic in cell fate decisions, we constructed a logic-incorporated GRN model and examined its behavior under two distinct driving forces (noise-driven and signal-driven). Under the noise-driven mode, we distilled the relationship among fate bias, regulatory logic, and noise profile. Under the signal-driven mode, we bridged regulatory logic and progression-accuracy trade-off, and uncovered distinctive trajectories of reprogramming influenced by logic motifs. In differentiation, we characterized a special logic-dependent priming stage by the solution landscape. Finally, we applied our findings to decipher three biological instances: hematopoiesis, embryogenesis, and trans-differentiation. Orthogonal to the classical analysis of expression profile, we harnessed noise patterns to construct the GRN corresponding to fate transition. Our work presents a generalizable framework for top-down fate-decision studies and a practical approach to the taxonomy of cell fate decisions.
-
- Developmental Biology
- Evolutionary Biology
Despite rapid evolution across eutherian mammals, the X-linked MIR-506 family miRNAs are located in a region flanked by two highly conserved protein-coding genes (SLITRK2 and FMR1) on the X chromosome. Intriguingly, these miRNAs are predominantly expressed in the testis, suggesting a potential role in spermatogenesis and male fertility. Here, we report that the X-linked MIR-506 family miRNAs were derived from the MER91C DNA transposons. Selective inactivation of individual miRNAs or clusters caused no discernible defects, but simultaneous ablation of five clusters containing 19 members of the MIR-506 family led to reduced male fertility in mice. Despite normal sperm counts, motility, and morphology, the KO sperm were less competitive than wild-type sperm when subjected to a polyandrous mating scheme. Transcriptomic and bioinformatic analyses revealed that these X-linked MIR-506 family miRNAs, in addition to targeting a set of conserved genes, have more targets that are critical for spermatogenesis and embryonic development during evolution. Our data suggest that the MIR-506 family miRNAs function to enhance sperm competitiveness and reproductive fitness of the male by finetuning gene expression during spermatogenesis.






