Developmental Neuroscience: Re-utilization of a transcription factor
A neuronal stem cell is a cell that divides to produce another neuronal stem cell plus a cell that can go on to become any one of the different types of neurons or glial cells that are found in the nervous system. Understanding how this happens is a major challenge in developmental neuroscience. In both vertebrates and invertebrates, the capacity of neuronal stem cells to produce specific cell types is determined by a combination of spatial patterning factors, essentially determined by their birth location, and temporal patterning, in which transcription factors that are expressed sequentially in the neuronal stem cells determine the kind of cells that are produced during a given time window (reviewed in Kohwi and Doe, 2013). Now, in eLife, Johannes Stratmann, Hugo Gabilondo, Jonathan Benito-Sipos and Stefan Thor – who are based at Linköping University and Universidad Autónoma de Madrid – report that Krüppel, a transcription factor that is involved in temporal patterning, has a bigger role than was previously realized (Stratmann et al., 2016).
In Drosophila, neuronal stem cells, known as neuroblasts, in the ventral nerve cord (Isshiki et al., 2001), the central brain (Bayraktar and Doe, 2013) and the optic lobes (Li et al., 2013) express a series of different temporal transcription factors as they divide, and this temporal patterning dictates the identity of the neurons that are produced over time. In the ventral nerve cord, Hunchback is expressed first, followed by Krüppel, Pdm, Castor and Grainyhead. Moreover, during the time window in which a given transcription factor is expressed, two or more different types of neurons can be produced. For example, at the end of the Castor window, a neuroblast called NB5-6 divides four times to sequentially produce four different types of neurons: Tv1, Tv2, Tv3 and Tv4 neurons.
The identity of each neuron formed during the Castor window is established by a complex web of transcriptional cascades that depend on Castor and other transcription factors (see Figure 1; Baumgardt et al., 2009; Baumgardt et al., 2007; Benito-Sipos et al., 2011). However, many of the details of the mechanisms that switch these cascades on and off at different times are unknown. Stratmann et al. shed light on the subject by looking at the role of Krüppel (Kr). The neuroblast being studied will already have passed through the Kr temporal window, but this transcription factor is expressed again in the neurons that are produced during the Castor window.
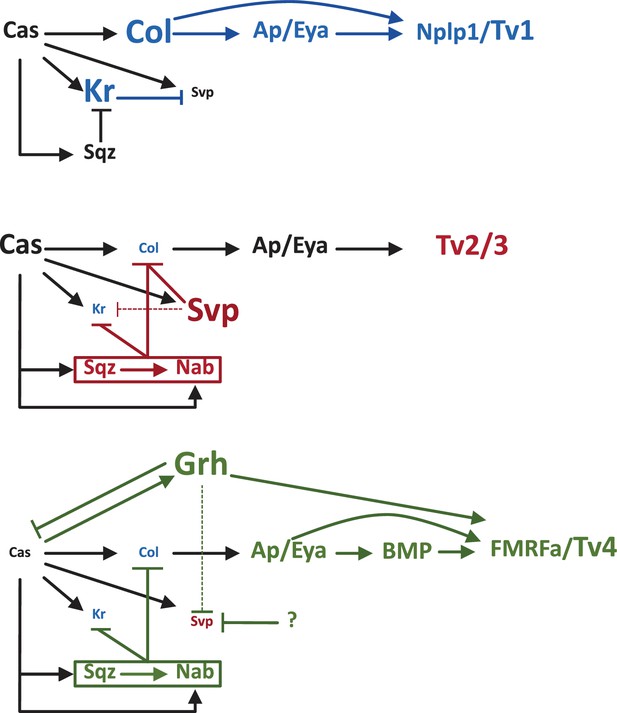
Establishing four different types of neurons in the Castor window.
Castor (Cas) is a temporal transcription factor expressed in NB5-6 neuroblasts. It activates the transcription factor Collier (Col), which then promotes the expression of Apterous and Eyes absent (Ap/Eya). In Tv1, Col and Ap/Eya form a feedforward loop to activate the neuropeptide called Nlpl1. This results in the neuroblast producing a Tv1 neuron (top). In Tv2/3 neurons, Cas activates a feedforward loop consisting of Squeeze (Sqz), Nab and Seven up (Svp) that inhibits Col, blocking the feedforward loop that leads to Nplp1 expression and allowing the establishment of the Tv2/3 identity (middle). In Tv4, Cas promotes the gradual expression of the next temporal transcription factor, Grainyhead (Grh), which inhibits Cas and Svp (thus blocking Tv2/Tv3 specification), while allowing the formation of a feedforward loop involving Ap/Eya and BMP that promotes the expression of the neuropeptide FMRFa. The neuroblast now produces a Tv4 neuron. Stratmann et al. explored the role of another temporal transcription factor, Krüppel (Kr), during the Cas window. They found that, at first, Kr inhibits Svp: this allows the establishment of the Col>Ap/Eya>Nplp1 feedforward loop that leads to the formation of Tv1 neurons (top; key players shown in blue). Later in the window, the Cas>Sqz>Nab feedforward loop inhibits both Kr and Col, as does Svp, which allows Tv2 and Tv3 neurons to form (middle; key players shown in red). Later in the window, a putative factor helps Grh to repress Svp, Kr remains inhibited by Sqz and Nab, and Grh promotes the expression of FMRFa that specifies Tv4 (bottom; key players shown in green). BMP: bone morphogenetic protein.
The default fate for neuroblasts at the end of the Castor window is to produce Tv2/3 neurons. However, Stratmann et al. show that Castor first activates Kr expression specifically in Tv1 neurons, which in turn inhibits expression of the nuclear hormone receptor Seven up (Svp). This receptor normally inhibits the expression of a transcription factor called Collier: however, the inhibition of Svp allows for the establishment of a feedforward loop downstream of Collier that leads to the expression of the neuropeptide Nplp1 and the establishment of Tv1 neuron identity (Figure 1).
As the neuroblast divides to produce Tv2 and Tv3, Svp and two other proteins – Squeeze (Sqz) and Nab – are expressed: this all leads to the inhibition of Kr and the repression of Collier. As a consequence the Nplp1 feedforward loop is inhibited and the default Ap/Eya identity in Tv2/3 is established. In the last division, as the neuroblasts gets ready to switch to the next temporal window, the increased expression of Grainyhead (the next temporal transcription factor in the series) has two effects: it promotes the expression of a neuropeptide called FMRFa, and it helps to inhibit Svp expression. The end result is to prevent the establishment of the generic Tv2/3 fate to promote the Tv4 identity. However, Grainyhead cannot inhibit Svp on its own, so another factor must also be expressed during this period to help it repress Svp (Figure 1).
The implications of these observations extend well beyond the details of the NB5-6 lineage and raise additional questions: How is Kr and Sqz expression scheduled? And how is Nab expression delayed to only be present in Tv1? Such delay could be explained by the formation of a feedforward loop in which Castor and Sqz combine to promote Nab expression (Baumgardt et al., 2009). The fact that Cas is only expressed at relatively low levels in Tv1 could also explain the absence of Nab expression in this neuron. Understanding such timing mechanisms will define the logic of this gene network, from the early expression of spatial and temporal transcription factors in the neuroblasts to the expression of the terminal differentiation genes that assign unique identities to neurons.
Many of the factors that are active during the Castor window are also active during other windows. As we have seen, Krüppel is active in neuroblasts during the Kr window and is active again in Tv1 neurons produced during the Castor window. Sub-temporal factors can also have multiple roles: Svp, for example, is expressed early in neuroblasts to regulate the transition from the Hunchback window to the Krüppel window (Kanai et al., 2005), and is also expressed in Tv neurons produced during the Castor window. This raises the question of how common such dual roles are among temporal genes.
The expression of Collier requires the input of several spatial transcription factors (Gabilondo et al., 2016; Karlsson et al., 2010), but this is not the case for the temporal factors Castor and Grainyhead and for the sub-temporal factors (Kr, Sqz, Nab and Svp) downstream of Castor. This suggests that the activation of the feedforward loops, but not of temporal factors, can depend on the activity of spatial factors expressed earlier during neuroblast formation. It will be interesting to test how the earlier expression of spatial factors modulates the DNA landscape of neuroblasts, and their progeny neurons, to control their competence to respond to the activity of temporal and sub-temporal transcription factors. If we can find the answers to such fundamental questions, it will improve our ability to produce specific neurons by controlling the fate of neural stem cells.
References
-
Temporal fate specification and neural progenitor competence during developmentNature Reviews Neuroscience 14:823–838.https://doi.org/10.1038/nrn3618
Article and author information
Author details
Publication history
- Version of Record published: October 14, 2016 (version 1)
Copyright
© 2016, Pinto-Teixeira et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,405
- Page views
-
- 131
- Downloads
-
- 2
- Citations
Article citation count generated by polling the highest count across the following sources: Crossref, PubMed Central, Scopus.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Developmental Biology
- Neuroscience
Human fetal development has been associated with brain health at later stages. It is unknown whether growth in utero, as indexed by birth weight (BW), relates consistently to lifespan brain characteristics and changes, and to what extent these influences are of a genetic or environmental nature. Here we show remarkably stable and lifelong positive associations between BW and cortical surface area and volume across and within developmental, aging and lifespan longitudinal samples (N = 5794, 4–82 y of age, w/386 monozygotic twins, followed for up to 8.3 y w/12,088 brain MRIs). In contrast, no consistent effect of BW on brain changes was observed. Partly environmental effects were indicated by analysis of twin BW discordance. In conclusion, the influence of prenatal growth on cortical topography is stable and reliable through the lifespan. This early-life factor appears to influence the brain by association of brain reserve, rather than brain maintenance. Thus, fetal influences appear omnipresent in the spacetime of the human brain throughout the human lifespan. Optimizing fetal growth may increase brain reserve for life, also in aging.
-
- Developmental Biology
- Immunology and Inflammation
During embryogenesis, the fetal liver becomes the main hematopoietic organ, where stem and progenitor cells as well as immature and mature immune cells form an intricate cellular network. Hematopoietic stem cells (HSCs) reside in a specialized niche, which is essential for their proliferation and differentiation. However, the cellular and molecular determinants contributing to this fetal HSC niche remain largely unknown. Macrophages are the first differentiated hematopoietic cells found in the developing liver, where they are important for fetal erythropoiesis by promoting erythrocyte maturation and phagocytosing expelled nuclei. Yet, whether macrophages play a role in fetal hematopoiesis beyond serving as a niche for maturing erythroblasts remains elusive. Here, we investigate the heterogeneity of macrophage populations in the murine fetal liver to define their specific roles during hematopoiesis. Using a single-cell omics approach combined with spatial proteomics and genetic fate-mapping models, we found that fetal liver macrophages cluster into distinct yolk sac-derived subpopulations and that long-term HSCs are interacting preferentially with one of the macrophage subpopulations. Fetal livers lacking macrophages show a delay in erythropoiesis and have an increased number of granulocytes, which can be attributed to transcriptional reprogramming and altered differentiation potential of long-term HSCs. Together, our data provide a detailed map of fetal liver macrophage subpopulations and implicate macrophages as part of the fetal HSC niche.






