Proteopedia is a tool for sharing knowledge about the structure and function of proteins, DNA, and other macromolecules on the web, developed by scientists at the Weizmann Institute of Science in Israel.
Blog post by Jaime Prilusky and Joel L. Sussman, from the Weizmann Institute of Science, Israel
Online publishing opens new opportunities for conveying scientific ideas in new ways. For example, Proteopedia makes it possible to publish ideas about the relationship between the structures of 3D biological macromolecules and their functions in novel, visual ways.
Proteopedia is a web-based Wiki for crowd sourcing concepts on the 3D structure/function relationships of biological macromolecules 1-4. As it ties text to 3D interactive images, Proteopedia aims to serve as a forum for the scientific community to share, retrieve, and discuss information related to proteins, macromolecules, small molecules, and chemicals of interest. It also aims to maintain low barriers to contribution.
These goals fit directly into how Proteopedia is currently being used in conjunction with publication of scientific papers and how it might be used in the future to aid more directly in the publication process.
There are already around 90 Interactive 3D Complements ( I3DC) in Proteopedia articles, which serve as complements to published papers in scientific journals. A link within the publication to an I3DC in Proteopedia enables readers to rotate molecular scenes with the same initial orientation, color schemes, and labeling as figures in the publication. Links to the I3DC in Proteopedia can be placed in the text of the publication, and/or in the online text file included on the journal website as supplementary material. The I3DC has a link directly back to the original journal article. It is not visible to the public until the online version of the article appears on the journal’s website.
All the I3DCs are listed at Proteopedia:I3DC. Below is an example of one I3DC, which is complementary to a paper in Cell: ‘ Structural linkage between ligand discrimination and receptor activation by type I interferons’ 5. By clicking the green links embedded in the text, the reader can change the position and representation on the 3D structure in the applet, obtaining a view that complements and enhances the concept described in words in the paper.
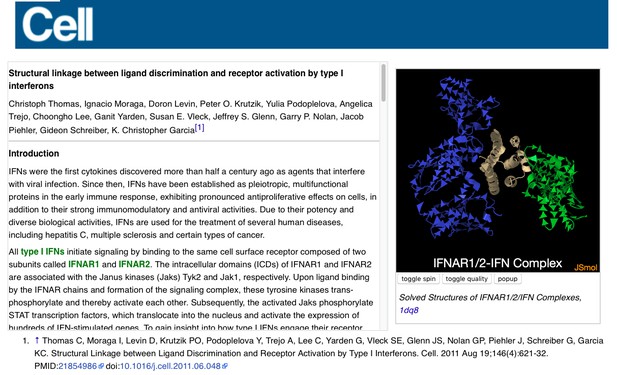
Example of an I3DC in Proteopedia, ‘ Structural linkage between ligand discrimination and receptor activation by type I interferons’, linking to a paper in Cell 5.
Although Proteopedia is a free web service, it differs from Wikipedia in that only registered users can contribute to it. The names of each contributor to a page in Proteopedia appear at the bottom of the page, giving them both credit and responsibility for what they write. At present, there are more than 3,300 registered users of Proteopedia from over 55 countries.
The text creation is done very simply via the use of MediaWiki Tools. The actual creation of the figures is via the user-friendly Graphical User Interface ‘ scene authoring tools’ (SATs) front end for the popular Jmol/JSmol web-based molecular visualization tool. In addition to the text describing the media wiki and SATs tools, there is a Proteopedia:Video Guide that contains seven short videos to help users quickly learn how to create Proteopedia pages.
Anyone, whether they are registered or not, can view pages in Proteopedia via computers, tablets or smartphones and see the 3D structures without needing any additional software beyond a standard internet browser.
In addition to around 120,000 automatically made pages in Proteopedia corresponding to every entry in the Protein Data Bank (PDB), there are about 3,000 user-added pages not directly tied to a particular published paper or particular entries in the PDB, but written more in the spirit of News & Views, describing a specific concept. Examples of a few of these pages can be seen below.
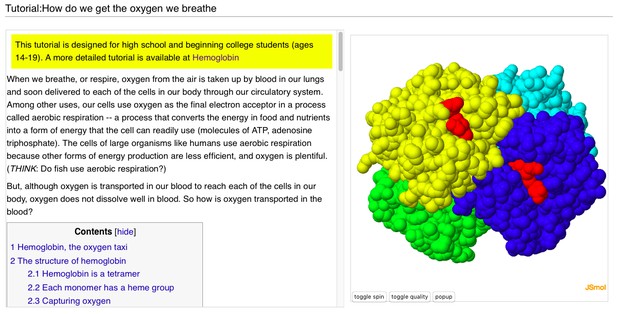
A user-contributed page in Proteopedia: ‘ Tutorial:How do we get the oxygen we breathe’.
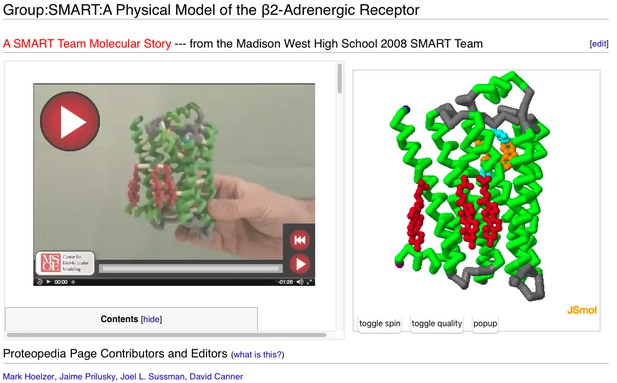
A user-contributed page in Proteopedia: ‘ Physical Model of the β2-Adrenergic Receptor’.
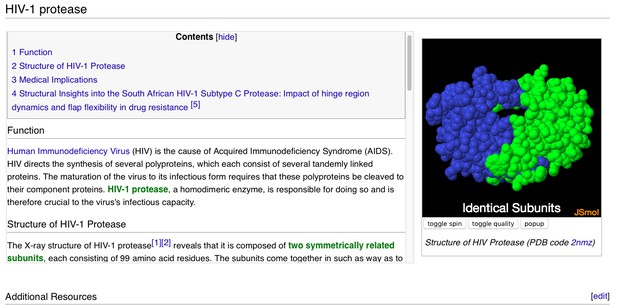
A user-contributed page in Proteopedia: ‘ HIV-1 Protease’.
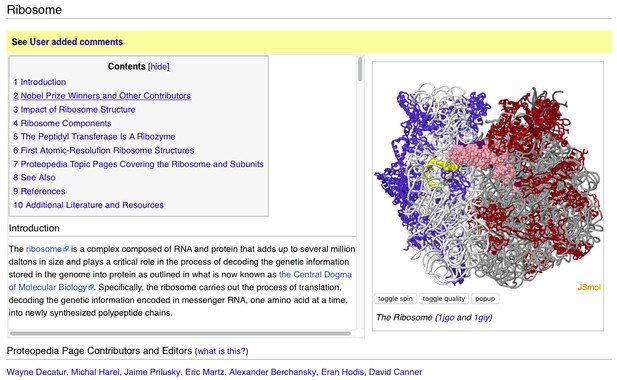
A user-contributed page in Proteopedia: ‘ Ribosome’.
You can get a feeling for how easy it is to generate a 3D live scene on Proteopedia at http://proteopedia.org/w/Proteopedia:DIY:Scenes. Following the step-by-step instructions for the first time might take 10-15 minutes, and all you need is a modern web browser (no Java required).
Over 3,300 users worldwide, including students at university and high school, are using Proteopedia to highlight structural aspects of proteins and their relation to function. We encourage you to give it a try.
References:
- Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. & Sussman, J. L. JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Israel J. Chem. 53, 207-216 (2013). http://dx.doi.org/10.1002/ijch.201300024
- Prilusky, J., Hodis, E. & Sussman, J. L. in Macromolecular Crystallography: Deciphering the Structure, Function and Dynamics of Biological Molecules Vol. XIV NATO Science for Peace and Security Series A: Chemistry and Biology (eds M.A. Carrondo & P. Spadon) 149-161 (Springer, 2012).
- Prilusky, J., Hodis, E., Canner, D., Decatur, W. A., Oberholser, K., Martz, E., Berchanski, A., Harel, M. & Sussman, J. L. Proteopedia: A status report on the collaborative, 3D web-encyclopedia of proteins and other biomolecules. J. Struct. Biol. 175, 244-252 (2011). http://dx.doi.org/10.1016/j.jsb.2011.04.011
- Hodis, E., Prilusky, J., Martz, E., Silman, I., Moult, J. & Sussman, J. L. Proteopedia – a scientific ‘wiki’ bridging the rift between 3D structure and function of biomacromolecules. Genome Biol. 9, R121 (2008). http://dx.doi.org/10.1002/bip.21126
- Thomas, C., Moraga, I., Levin, D., Krutzik, P. O., Podoplelova, Y., Trejo, A., Lee, C., Yarden, G., Vleck, S., Glenn, J. S., Nolan, G. P., Piehler, J., Schreiber, G. & Garcia, K. C. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146, 621-632 (2011). http://dx.doi.org/10.1016/j.cell.2011.06.04




