Deep evolutionary origin of gamete-directed zygote activation by KNOX/BELL transcription factors in green plants
Abstract
KNOX and BELL transcription factors regulate distinct steps of diploid development in plants. In the green alga Chlamydomonas reinhardtii, KNOX and BELL proteins are inherited by gametes of the opposite mating types and heterodimerize in zygotes to activate diploid development. By contrast, in land plants such as Physcomitrium patens and Arabidopsis thaliana, KNOX and BELL proteins function in sporophyte and spore formation, meristem maintenance and organogenesis during the later stages of diploid development. However, whether the contrasting functions of KNOX and BELL were acquired independently in algae and land plants is currently unknown. Here, we show that in the basal land plant species Marchantia polymorpha, gamete-expressed KNOX and BELL are required to initiate zygotic development by promoting nuclear fusion in a manner strikingly similar to that in C. reinhardtii. Our results indicate that zygote activation is the ancestral role of KNOX/BELL transcription factors, which shifted toward meristem maintenance as land plants evolved.
Introduction
The life cycles of eukaryotes alternate between diploid (2n) and haploid (n) phases through meiosis and fertilization (Bowman et al., 2016). In land plants, both the haploid and diploid phases are multicellular, producing gametophytic and sporophytic bodies, respectively. In bryophytes including liverworts, mosses, and hornworts, gametophytes are larger and more morphologically complex than sporophytes, which consist of only a few cell types. During the course of land plant evolution, the life cycle shifted toward a sporophyte-dominant style, presumably to facilitate adaptation to terrestrial environments where it is advantageous to generate large sporophytes that produce many spores. Consequently, the sporophytes of extant flowering plants (angiosperms) exhibit far more complex morphologies than their male and female gametophytes, the pollen grain, and embryo sac, respectively, which are composed of only a few cells. This evolutionary transition in life cycle is thought to have been facilitated by the cooption of genes and/or gene regulatory networks that regulate gametophyte development to function in sporophyte development (Bowman et al., 2019). However, key steps of life cycle progression per se have continued to be driven by conserved regulators during land plant evolution, as recently reported for gametophytic sexual differentiation and gamete formation (Koi et al., 2016; Rövekamp et al., 2016, Higo et al., 2018; Yamaoka et al., 2018; Hisanaga et al., 2019a; Hisanaga et al., 2019b).
Homeodomain transcription factors (HD TFs) are developmental regulators that are evolutionarily conserved in eukaryotes. HD TFs are classified into two families, three-amino-acid-loop-extension (TALE) and non-TALE, based on amino acid sequence similarity in the HD domain (Bertolino et al., 1995; Derelle et al., 2007). In the fungi Saccharomyces cerevisiae and Coprinopsis cinerea, TALE and non-TALE HD TFs are expressed in haploid cells of opposite mating types (Herskowitz, 1989; Kues et al., 1992). These proteins heterodimerize in zygotes to regulate the expression of genes promoting the haloid-to-diploid transition (Goutte and Johnson, 1988; Spit et al., 1998). In green plants, TALE HD TFs have diversified into the KNOX (KNOTTED1-LIKE HOMEOBOX) and BELL (BELL-LIKE) subfamilies. In the unicellular green alga Chlamydomonas reinhardtii, the KNOX protein GAMETE SPECIFIC MINUS1 (GSM1) and the BELL protein GAMETE SPECIFIC PLUS1 (GSP1) accumulate in the cytosol of minus and plus gametes, respectively. Upon fertilization, the two proteins heterodimerize and translocate to both male and female pronuclei to activate the expression of early zygote-specific genes. Loss-of-function mutations in either GSP1 or GSM1 result in pleiotropic phenotypes involving cellular rearrangements in zygotes, such as the loss of nuclear and mitochondrial fusion, lack of selective degradation of minus-derived chloroplast DNA and chloroplast membrane fusion, and defects in flagellar resorption (Joo et al., 2017; Kariyawasam et al., 2019; Lee et al., 2008; Lopez et al., 2015; Nishimura et al., 2012).
In land plants, KNOX proteins are further diversified into class I (KNOX1) and class II (KNOX2) (Kerstetter et al., 1994; Mukherjee et al., 2009). The developmental functions of these proteins have been studied extensively in angiosperms such as maize (Zea mays), rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana) (reviewed in Hay and Tsiantis, 2010). Based on their expression patterns and the phenotypes of both knockout and overexpression lines, KNOX1 proteins are thought to promote cell proliferation in the meristematic tissues of aerial organs. The biological functions of KNOX2 genes are somewhat elusive, but they are thought to act antagonistically to KNOX1 to promote cell differentiation (Furumizu et al., 2015).
The apparent functional dissimilarity of KNOX proteins between C. reinhardtii and Arabidopsis (zygote activation versus cellular proliferation/differentiation) may reflect the large phylogenetic distance between these two species as they separated into two major green plant lineages, Chlorophyta and Streptophyta, some 700 million years ago (Becker, 2013). Functional analyses of KNOX genes in the moss Physcomitrium patens, however, pointed to some commonality between the two KNOX functions in sporophyte development. The moss genome contains three KNOX1 and two KNOX2 genes, which are all primarily expressed in sporophytes, though expression of one KNOX1 and two KNOX2 genes is additionally detected in egg cells (Horst et al., 2016; Sakakibara et al., 2013; Sakakibara et al., 2008). A triple loss-of-function mutant of all three KNOX1 genes was defective in cell division and differentiation in sporophytes, as well as spore formation (Sakakibara et al., 2008). By contrast, simultaneous knockout of the two KNOX2 genes resulted in ectopic gametophyte formation in sporophyte bodies (Sakakibara et al., 2013). Thus, at least in one bryophyte species, KNOX1 and KNOX2 control sporophyte development via two pathways, with one ensuring proper sporophyte development (like C. reinhardtii GSM1) and the other promoting cell proliferation (like Arabidopsis KNOX1 proteins). While the transition of the role of KNOX/BELL from zygote activation to sporophyte morphogenesis likely arose during plant evolution, the point in plant phylogeny at which this transition occurred is unclear.
Here, we analyzed the roles of KNOX1 and BELL in the liverwort Marchantia polymorpha, a model species suitable to study evolution of sexual reproduction in plants (Hisanaga et al., 2019b). We uncovered unexpected conservation of KNOX/BELL function between the phylogenetically distant green plants M. polymorpha and C. reinhardtii, but not between the more closely related M. polymorpha and P. patens. Thus, the functional transition of KNOX/BELL from zygote activation to sporophyte morphogenesis occurred at least once in the land plant lineage independently of the acquisition of multicellular sporophytes. Additionally, we uncovered inverted sex-specific expression patterns of KNOX and BELL genes between C. reinhardtii and M. polymorpha, suggesting that anisogamy evolved independently of KNOX/BELL expression in gametes.
Results
MpKNOX1 is an egg-specific gene in M. polymorpha
We previously reported that an RWP-RK TF MpRKD promotes egg cell differentiation in M. polymorpha. Loss-of-function Mprkd mutant females grow normally and produce archegonia like the wild-type, but their egg cells do not mature, instead degenerating after ectopic cell division and vacuolization (Koi et al., 2016). We made use of this egg-specific defect in Mprkd to identify genes preferentially expressed in egg cells of M. polymorpha. Briefly, we collected ~2000 archegonia from two independent Mprkd female mutant lines (Mprkd-1 and Mprkd-3; Koi et al., 2016), each in two replicates. As a control, ~4000 archegonia were collected from wild-type females in four replicates. We extracted RNA from each pool and analyzed it by next-generation sequencing. Comparative transcriptome analysis identified 1583 and 170 genes with significantly decreased and increased mRNA levels, respectively, in Mprkd compared to wild-type archegonia, respectively (≥3-fold and false discovery rate <0.01; Figure 1A and Figure 1—figure supplement 1A, Supplementary file 1). Among the genes with reduced expression levels, MpKNOX1 (Mp5g01600), the only class I KNOX gene in M. polymorpha (Bowman et al., 2017, Frangedakis et al., 2017), showed more than 300-fold reduced expression level in Mprkd as compared to that in the wild-type (fragments per kilobase of exon per million mapped reads (FPKM) values are 19.5, 0.0629, and 0.0124 in wild-type, Mprkd-1, and Mprkd-3 archegonia, respectively; Supplementary file 1). The MpKNOX1 polypeptide contains KNOX I, KNOX II, ELK, and Homeobox domains, as do KNOX proteins from green algae, mosses, ferns, and flowering plants (Figure 1B).
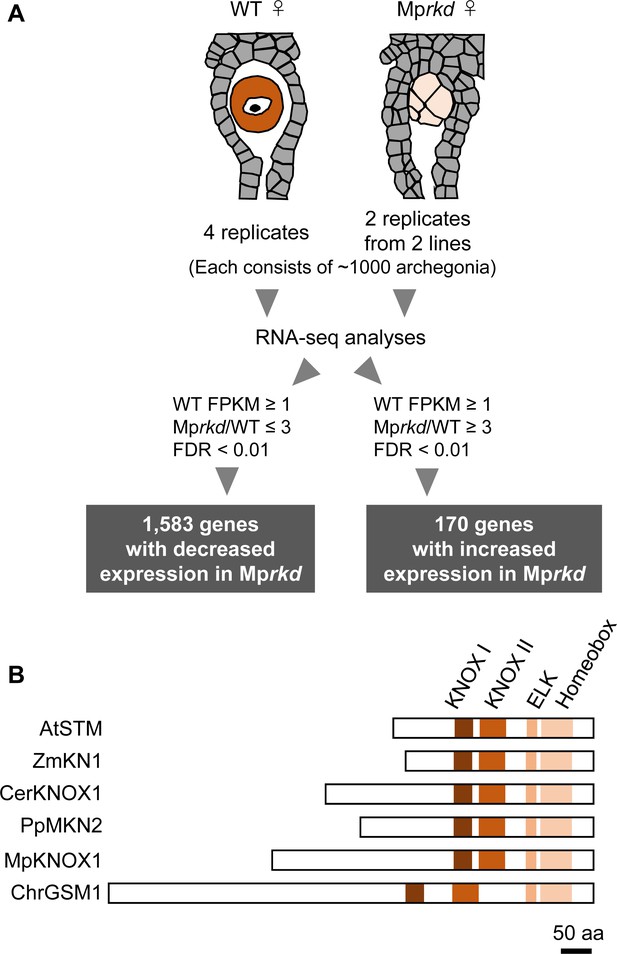
Comparative transcriptome analysis of wild-type and Mprkd archegonia and identification of MpKNOX1 as an egg-specific gene.
(A) Schematic illustration of RNA-seq analysis comparing the archegonia transcriptomes from wild-type females and egg-deficient Mprkd mutant females. About 4000 and 2000 archegonia were collected from wild-type and each of the two Mprkd mutant lines, and randomly allocated into four and two replicates, respectively. (B) Comparison of the domain arrangements of MpKNOX1 vs. representative class I KNOX proteins from Arabidopsis thaliana (AtSTM; GenBank accession number AEE33958.1), Zea mays (ZmKN1; AAP21616.1), Ceratopteris richardii (CerKNOX1, BAB18582.1), Physcomitrium patens (PpMKN2; AAK61308.2), and Chlamydomonas reinhardtii (ChrGSM1; ABJ15867.1).
Previous RNA-sequencing data (Bowman et al., 2017) indicated that MpKNOX1 is specifically expressed in female plants (Figure 1—figure supplement 1B). To obtain the detailed expression patterns of MpKNOX1, we performed RT-PCR analysis using RNA extracted from vegetative and reproductive organs of male and female gametophytes, as well as 3-week-old sporophytes, and confirmed the specific expression of MpKNOX1 in archegoniophores (Figure 2A). No expression was detected in female or male thalli (leaf-like vegetative organs), antheridiophores (male reproductive branches), or sporophytes (Figure 2A). To visualize the cell- and tissue-specific expression patterns of MpKNOX1, we generated a MpKNOX1 transcriptional reporter line (MpKNOX1pro:H2B-GFP). Consistent with the >300-fold reduced MpKNOX1 transcript levels in egg-deficient Mprkd archegonia (Figure 1B), reporter expression was specifically detected in egg cells (Figure 2C) and not in developing archegonia before the formation of egg progenitors (Figure 2B). Together, these data indicate that MpKNOX1 is an egg-specific gene in M. polymorpha.
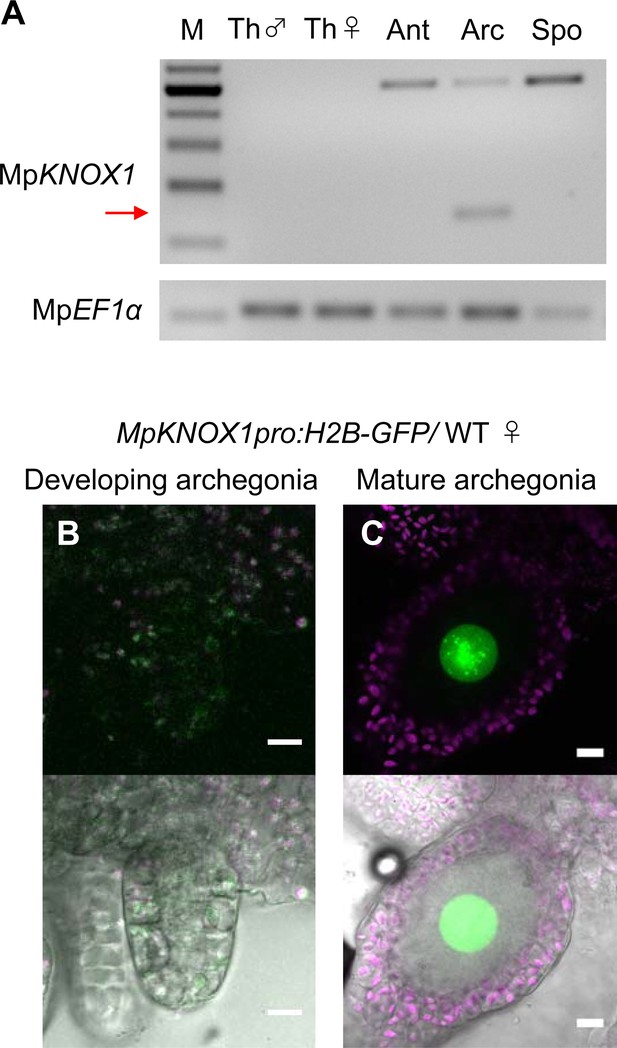
MpKNOX1 is specifically expressed in egg cells.
(A) RT-PCR analysis of MpKNOX1. Lanes are labeled as follows: M: size markers; Th ♂: male thalli; Th ♀: female thalli; Ant: antheridiophores; Arc: archegoniophores; Spo: sporophytes of 3-week-old plants. Constitutively expressed MpEF1α was used as a control. Red arrow indicates the expected size of PCR products from spliced MpKNOX1 mRNA. Bands at the top of the gel likely correspond to unspliced MpKNOX1 transcripts. Shown is a representative result from the experiments using three independently collected plant samples each with two technical replicates (two PCRs from each cDNA pool). See Figure 4—figure supplement 1A for the primer position. (B, C) Expression of the MpKNOX1 transcriptional reporter. Magenta: chlorophyll autofluorescence; green: GFP fluorescence. Lower panels are merged photographs of fluorescence and bright-field images. Bars, 10 μm.
Zygote activation of wild-type M. polymorpha lags during karyogamy
The egg-specific expression of MpKNOX1 attracted our attention as this pattern is in contrast to the previously reported function of KNOX1s in sporophyte morphogenesis in land plants (Furumizu et al., 2015; Sakakibara et al., 2008). Instead, the egg-specific expression of MpKNOX1 is reminiscent of GSM1, a KNOX homolog in C. reinhardtii that is expressed in plus gametes and activates zygote development after fertilization (Joo et al., 2017; Kariyawasam et al., 2019; Lee et al., 2008; Lopez et al., 2015; Ning et al., 2013; Nishimura et al., 2012). Therefore, we explored whether MpKNOX1 plays a role in zygote activation in M. polymorpha.
While the processes of gametogenesis and embryo patterning in M. polymorpha (Figure 3A) have been characterized histologically (Durand, 1908; Higo et al., 2016; Koi et al., 2016; Shimamura, 2016; Zinsmeister and Carothers, 1974), the subcellular dynamics associated with zygote activation have not been described in detail. To visualize these dynamics, we established a simple in vitro fertilization method for M. polymorpha. Briefly, several antheridiophores and archegoniophores were co-cultured for 1 hr in a plastic tube containing an aliquot of water to allow sperm to be released from the antheridia and enter into the archegonia (Figure 3B). At the end of the co-culture period, sperm nuclei were visible in most eggs (Figure 3C), indicating that fertilization had been completed during the 1 hr of co-culturing. Subsequently, sperm-containing archegonia were transferred to a fresh tube containing water and cultured for 2 weeks (Figure 3B). This experimental regime restricted the timing of sperm entry to the 1 hr time window of the co-culture period (whose termination is defined hereafter as the time of fertilization), allowing us to perform a time-course observation of fertilization and embryogenesis (Figure 3D–I).
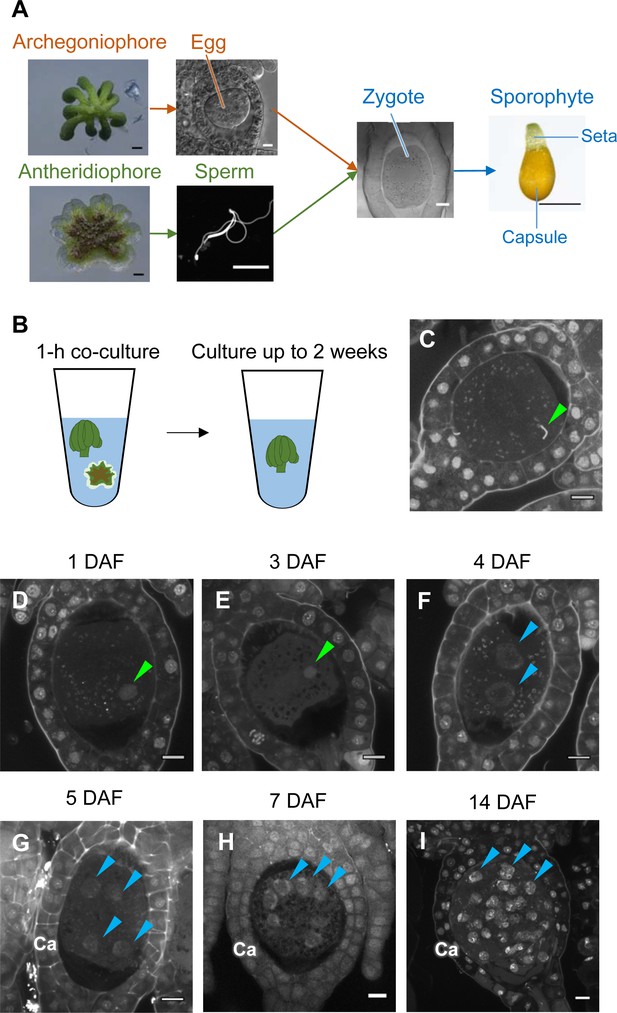
Time-course observation of subcellular dynamics during M. polymorpha fertilization.
(A) Schematic representation of sexual reproduction in M. polymorpha. Female plants develop umbrella-shaped sexual branches termed archegoniophores that form egg-containing archegonia. Male plants develop disc-shaped sexual branches termed antheridiophores that form antheridia, which produce numerous motile sperm cells. Upon soaking antheridiophores in water, the sperm cells are released from the antheridia and swim to egg cells in the archegonia. After fertilization, each zygote undergoes embryogenesis by dividing and differentiating into a sporophyte body consisting of a capsule containing haploid spores and a short supportive stalk called the seta. Black bars, 1 mm. White bars, 10 µm. (B) Illustration of the in vitro fertilization method used in this study. Excised archegoniophores and antheridiophores were co-cultured in water for 1 hr to allow fertilization to take place. The archegoniophores were transferred to a fresh tube containing water for further culturing. The tube lids were left open to allow gas exchange to occur. Archegoniophores containing sporophytes were cultured for up to 2 weeks. (C) A DAPI-stained zygote after 1 hr of co-culture. Most zygotes contained sperm nuclei at this time (green arrowhead). Bars, 10 µm. (D–I) DAPI-stained zygotes and sporophytes at the indicated days after fertilization (DAF). Male pronuclei (green arrowheads) were visible at 1–3 DAF (D, E) in wild-type fertilized eggs. In most zygotes, karyogamy was completed, and cells were cleaved at 4 DAF (F). Sporophyte cells continued to divide at 5–14 DAF (G–I), as visualized by the presence of multiple nuclei (blue arrowheads; not all nuclei are labeled in H and I). Ca: calyptra. Bars, 10 µm.
We observed the cellular dynamics of zygotes and early embryos using optimized cell wall staining and tissue clearing techniques (Miyashima et al., 2019; Kurihara et al., 2015). No cell walls were stained in mature egg cells (Figure 3—figure supplement 1A), indicating that cell walls are not present in mature eggs, a prerequisite for fusion with sperm cells. A cell wall around the zygote was detected only at 3 days after fertilization (DAF) (Figure 3—figure supplement 1B–D), whereas the first zygotic division occurred at 4–5 DAF (Figure 3—figure supplement 1E and F). At 1–3 DAF, a male pronucleus was clearly stained with DAPI, in contrast to the female pronucleus (not visible by DAPI staining), and was typically positioned halfway between the periphery and center of the fertilized egg (Figure 3D and E, green arrowhead). At 4 DAF, most zygotes completed the first division (Figure 3F), indicating that karyogamy takes place at 3–4 DAF. After 5 DAF, zygotes and the surrounding archegonial wall cells divided to form sporophytes and the calyptra (a protective gametophyte tissue), respectively (Figure 3G–I). These cellular rearrangements, including karyogamy and embryogenesis, proceeded at a rate comparable to that in zygotes produced in planta (Figure 3—figure supplement 2), confirming that our in vitro fertilization protocol faithfully recapitulated fertilization programs in planta.
Maternal MpKNOX1 is required for pronuclear fusion in zygotes
To analyze the biological functions of MpKNOX1, we generated loss-of-function mutants of MpKNOX1 using a CRISPR/Cas9 technique optimized for M. polymorpha (Sugano et al., 2018). We obtained three independent female mutant lines that harbored nucleotide insertions or deletions resulting in premature stop codons preceding the region encoding the HD (Figure 4—figure supplement 1A). All mutants were indistinguishable from wild-type females in terms of both vegetative and reproductive morphology (Figure 4—figure supplement 2). Mature archegonia and egg cells of the Mpknox1 mutants were also indistinguishable from those of the wild-type (Figure 4A and E), indicating that MpKNOX1 functions are dispensable for both gametophyte development and gametogenesis.
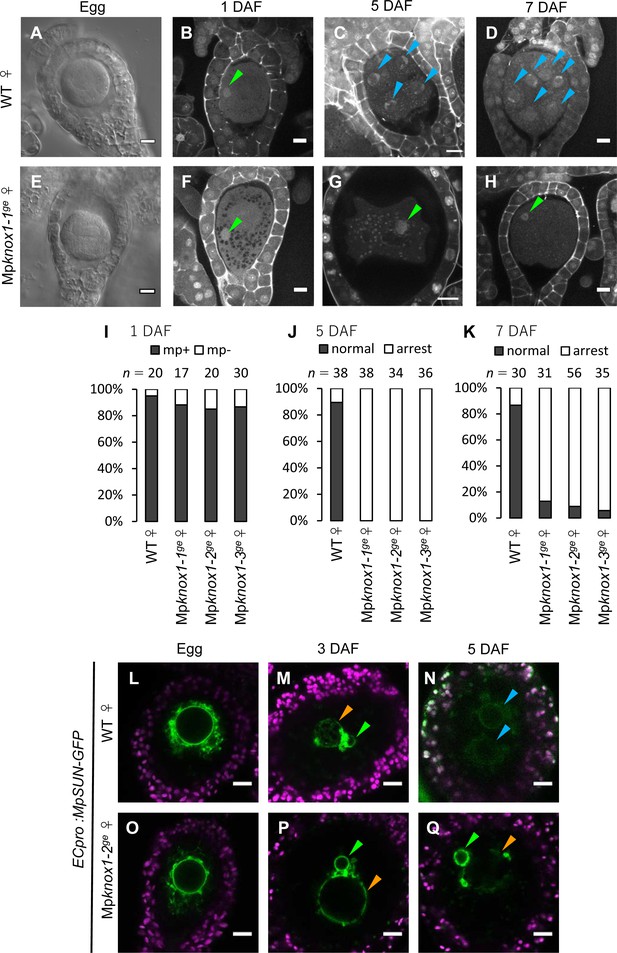
Maternally inherited MpKNOX1 is required for nuclear fusion.
(A, E) Bright-field images of wild-type (WT; A) and Mpknox1-1ge (E) archegonia. (B–D and F–H). 1 day after fertilization (DAF) (B, F), 5 DAF (C, G), and 7 DAF (D, H) zygotes from a cross between WT female and male plants (B–D), and a cross between Mpknox1-1ge female and WT male plants (F–H), indicating that maternal MpKNOX1 is dispensable for fertilization (B, F) but is required for embryogenesis (C, D, G, H). (I–K) Bar graphs showing the ratios of zygotes containing male pronuclei (mp+) vs. those not containing male pronuclei (mp-) at 1 DAF (I), and developed vs. arrested zygotes at 5 DAF (J) and 7 DAF (K) derived from the crosses of either WT or Mpknox1 females with WT males. Number of observed zygotes is shown above each bar. (L–Q) Egg cells of MpSUN-GFP marker lines in the WT (L) or Mpknox1-2ge (O) female background were crossed with WT males. At 3 DAF, male and female pronuclei were in contact with each other in both WT (M) and Mpknox1 (P) eggs. At 5 DAF, zygotes derived from a WT egg started to divide (N), while those from an Mpknox1 female (Q) were arrested without nuclear membrane fusion. Green arrowhead: male pronucleus; orange arrowhead: female pronucleus; blue arrowhead: embryo nucleus. Bars, 10 µm.
We crossed the Mpknox1 mutant females with wild-type males and observed the resulting zygotes by microcopy. Similar to wild-type zygotes, each Mpknox1 egg fertilized with wild-type sperm harbored a male pronucleus at 1 DAF (Figure 4B, F and I), indicating that Mpknox1 eggs are able to fuse with wild-type sperm and support decondensation of sperm nuclei. At 5 DAF, however, male and female pronuclei remained unfused in fertilized Mpknox1 eggs (100%, n = 34–38, Figure 4G and J), in contrast with wild-type fertilized eggs, a majority of which were undergoing sporophyte development (89%, n = 38, Figure 4C and J). At 7 DAF, most Mpknox1 eggs contained unfused male and female pronuclei (87–94%, n = 31–56, Figure 4H and K). The three independent Mpknox1 mutant lines exhibited indistinguishable defects in karyogamy and sporophyte development (Figure 4I–K). Importantly, these defects were rescued in archegonia expressing MpKNOX1-GFP by the MpKNOX1 promoter (gMpKNOX1-GFP, Figure 4—figure supplement 3), confirming the notion that MpKNOX1 is required maternally to complete fertilization.
A small fraction of Mpknox1 eggs fertilized with wild-type sperm developed into sporangia that produced functional spores (Figure 4—figure supplement 4), suggesting that redundant genetic pathway(s) can compensate for the loss of MpKNOX1 and/or that our Mpknox1 mutant alleles were not null, though genomic sequences preceding the HD-coding region were disrupted. This residual embryogenic capacity allowed us to obtain male Mpknox1 gametophytes from the resulting spores. The male Mpknox1 mutants produced functional sperm capable of producing normal embryos when used to fertilize wild-type eggs (Figure 4—figure supplement 5), indicating that paternal MpKNOX1 is dispensable for gametophyte development, fertilization, and embryogenesis. To further examine the lack of paternal contribution of MpKNOX1 to fertilization and sporophyte development, we crossed Mpknox1 females with Mpknox1 males. Resulting homozygous Mpknox1 zygotes exhibited the karyogamy and sporangia formation defects indistinguishable from those observed in zygotes carrying maternally inherited Mpknox1 alone (Figure 4—figure supplements 4 and 6). Together, these results indicate that the egg-derived functional MpKNOX1 allele or its protein products, but not those derived from sperm, are required to activate zygote development, more specifically karyogamy, during M. polymorpha fertilization.
In both animals and plants, karyogamy occurs via a two-step process: pronuclear migration and nuclear membrane fusion (Fatema et al., 2019). During pronuclear migration, one or both pronuclei migrate to become in close proximity, while during nuclear membrane fusion, the nuclear envelopes of the two pronuclei fuse together to produce a zygotic nucleus with both maternal and paternal genomes. To identify which of these steps is affected by the Mpknox1 mutation, we visualized the pronuclear envelope by expressing GFP-tagged Sad1/UNC84 (SUN) domain-containing proteins (MpSUN; Mp5g02400) under the control of an egg-specific promoter (ECpro:MpSUN-GFP, see Materials and methods for details). The SUN proteins are known to localize specifically to nuclear envelope in animals, yeasts, and vascular plants (Graumann et al., 2010; Tzur et al., 2006). Before fertilization, wild-type and Mpknox1 egg nuclei were of a similar size (approximately 20 µm in diameter) and were surrounded by a mesh-like membranous structure (Figure 4L and O). At 3 DAF, ECpro:MpSUN-GFP signals were visualized at the surfaces of both female and male pronuclei, suggesting the presence of a nuclear envelope (Figure 4M and P). In both wild-type- and Mpknox1-derived zygotes, female and male pronuclei were tethered to each other by a membranous structure marked by MpSUN-GFP (Figure 4M and P). At 5 DAF, however, male and female pronuclei remained separated by an intercalating membranous structure in Mpknox1-derived zygotes, while those from wild-type eggs already showed two embryonic nuclei (Figure 4N and Q). Together, these observations indicate that maternal MpKNOX1 or its protein product is dispensable for the organization and migration of pronuclei but is required for pronuclear membrane fusion.
Both maternal and paternal MpBELL alleles contribute to karyogamy
KNOX proteins heterodimerize with BELL proteins to regulate gene transcription (Hay and Tsiantis, 2010). In C. reinhardtii, a minus gamete-derived KNOX protein (GSM1) heterodimerizes with a plus gamete-derived BELL protein (GSP1) upon fertilization and activates the majority of early zygote-specific genes (Joo et al., 2017; Kariyawasam et al., 2019; Lee et al., 2008; Lopez et al., 2015; Nishimura et al., 2012). Loss of GSM1 and/or GSP1 results in pronuclear fusion arrest, a phenotype similar to that of Mpknox1 mutants. The phenotypic similarity between M. polymorpha Mpknox1 and C. reinhardtii gsm1 mutants suggests that the role of KNOX and BELL in zygote activation is conserved and that its evolutionary origin can be traced back to a common ancestor of the two species.
In support of this hypothesis, publicly available transcriptome data (Bowman et al., 2017) indicate that two of the five BELL genes of M. polymorpha, MpBELL3 (Mp8g02970) and MpBELL4 (Mp8g07680), are preferentially expressed in the antheridiophores of male plants, whereas MpBELL1 (Mp8g18310) and MpBELL5 (Mp5g11060) are preferentially expressed in sporophytes and archegonia, respectively (Figure 5—figure supplement 1). RT-PCR analysis confirmed that MpBELL3 and MpBELL4 are specifically expressed in antheridiophores containing antheridia (Figure 5A).
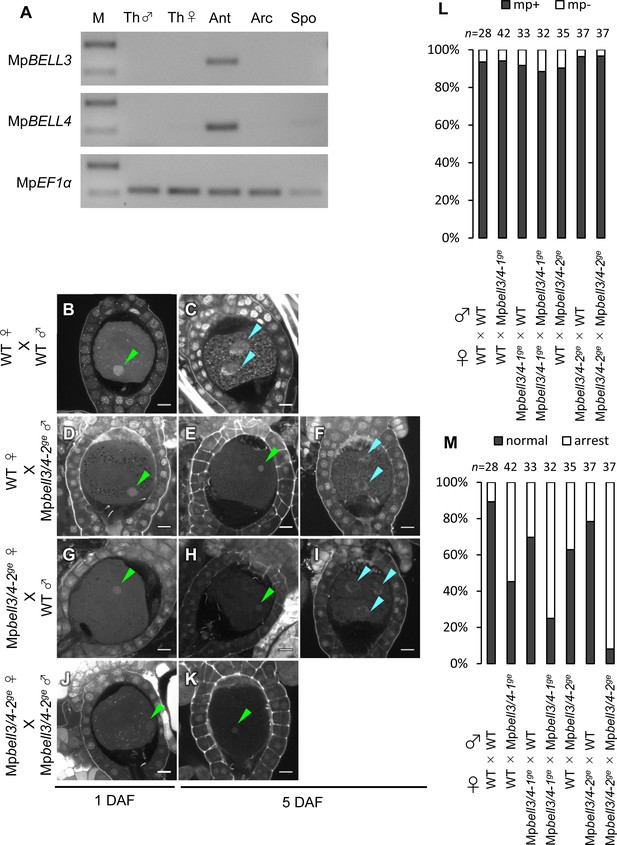
Both paternally and maternally inherited MpBELL genes are required for karyogamy.
(A) RT-PCR analysis indicating that MpBELL3 and MpBELL4 are specifically expressed in antheridiophores. The lanes are labeled as in Figure 2A. Shown is a representative result from the experiments using three independently collected samples each with two technical replicates. See Figure 4—figure supplement 1A for the primer position. (B–K) Zygotes at 1 day after fertilization (DAF; B, D, G, J) and 5 DAF (C, E, F, H, I, K) from crosses between a wild-type female and wild-type male (B, C) or Mpbell3/4-2ge male (D–F) and a Mpbell3/4-2ge female and a wild-type male (G–I) or Mpbell3/4-2ge male (J, K). The presence of male pronuclei (green arrowheads) in zygotes of all genotypes at 1 DAF (B, D, G, J) indicates that MpBELL3 and MpBELL4 are dispensable for plasmogamy. Note that zygotes produced from both or one Mpbell3/4 parent exhibit a variable degree of karyogamy arrest as visualized by the retention of male pronuclei (green arrowheads) among those starting embryonic division (nuclei labeled with blue arrowheads). Bars, 10 µm. (L) A bar graph showing the ratios of zygotes containing male pronuclei (mp+) vs. those not containing male pronuclei (mp-) from indicated crosses at 1 DAF. Numbers of observed zygotes are shown above each bar. (M) A bar graph showing the ratios of developed vs. arrested zygotes at 5 DAF from the indicated crosses. Numbers of observed zygotes are shown above each bar.
To investigate whether MpBELL3 and/or MpBELL4 are required for fertilization, we generated two male and one female mutant lines in which MpBELL3 and MpBELL4 were simultaneously disrupted (hereafter referred to as Mpbell3/4) using a CRISPR/Cas9 nickase system (Figure 4—figure supplement 1B and C). Both female and male Mpbell3/4 gametophytes grew normally (Figure 4—figure supplement 2) and produced normal gametangia and gametes capable of fertilization (Figure 5—figure supplement 2, Figure 5B, D, G, J and L). By contrast, approximately a half of zygotes produced by crossing wild-type females and Mpbell3/4 males did not complete karyogamy by 5 DAF (55% and 37% for Mpbell3/4-1ge and Mpbell3/4-2ge, respectively; Figure 5E, F and M).
Somewhat unexpectedly, zygotes obtained from a reciprocal cross (Mpbell3/4 females with wild-type males) exhibited a low but significant degree of karyogamy arrest (30% and 22% for Mpbell3/4-1ge andMpbell3/4-2ge, respectively; Figure 5H, I and M), despite the lack of detectable MpBELL3/4 expression in female gametes. Furthermore, a majority of 5-DAF zygotes obtained from a cross between Mpbell3/4 females and Mpbell3/4 males were arrested at karyogamy (75% and 92% for Mpbell3/4-1ge and Mpbell3/4-2ge, respectively; Figure 5K and M). A similar but slightly less proportion of arrested zygotes was observed at 7 DAF for each genotype (Figure 5—figure supplement 3A–G). Moreover, unfused pronuclei were not observed in any of the developing embryos harboring Mpbell3/4 allele(s) (Figure 5—figure supplement 3C and E). Consistently, the number of sporangia in Mpbell3/4 homozygotes was about a third of that in wild-type (Figure 5—figure supplement 3H). Taken together, these results suggest that not only paternally inherited MpBELL3 and/or MpBELL4 alleles(s) and/or their protein products, but also maternally inherited MpBELL3 and/or MpBELL4 allele(s) contribute to sporophyte development. A correlation between the proportion of arrested zygotes and the degree of reduction in sporangia number in each genotype suggests that MpBELL3/4 mainly act to promote karyogamy for the progression of sporophyte development.
MpKNOX1 proteins transiently localize to male and female pronuclei in an MpBELL3/4-dependent manner
Our genetic analyses indicated that MpKNOX1 functions after fertilization despite its specific transcription in unfertilized egg cells. This observation suggests that MpKNOX1 protein produced in unfertilized egg cells functions in zygotes. To test this hypothesis, we analyzed MpKNOX1 protein dynamics during fertilization by examining gMpKNOX1-GFP plants by confocal microscopy. In mature egg cells, MpKNOX1-GFP was specifically detected in the cytosol (Figure 6A). After crossing with a wild-type male, GFP signals were detected in both male and female pronuclei at 12 hours after fertilization (HAF) (83%, n = 48, Figure 6B and G). At 24 HAF, the GFP signals were totally excluded from the pronuclei (Figure 6C). These observations suggest that upon fertilization MpKNOX1 translocates from the cytosol to pronuclei.
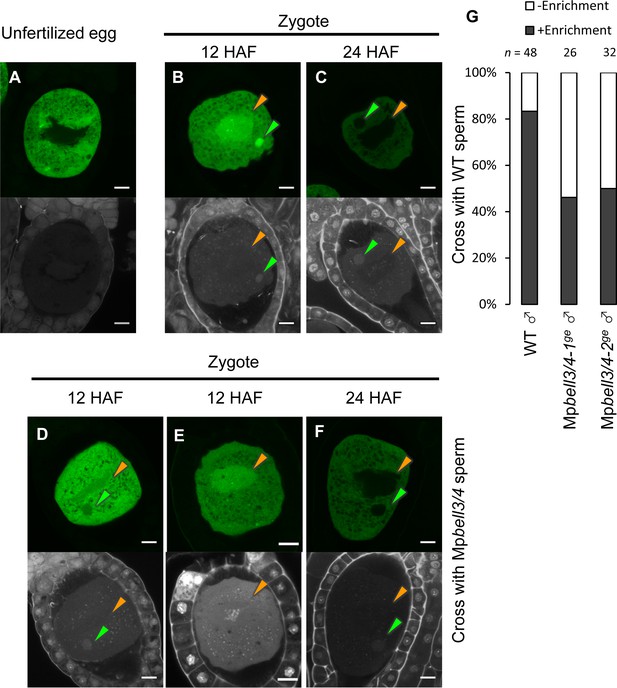
MpKNOX1 transiently localizes to female and male pronuclei prior to karyogamy.
(A–F) GFP (upper panels) and DAPI (lower panels) signals from gMpKNOX1-GFP/Mpknox1-1ge eggs (A) and zygotes obtained by crossing a gMpKNOX1-GFP/Mpknox1-1ge female with a wild-type (B, C) or Mpbell3/4-2ge (D–F) male. Note that before fertilization MpKNOX1-GFP was exclusively localized to the cytosol (A). At 12 HAF, MpKNOX1-GFP signals were enriched in female (orange arrowhead) and male (green arrowhead) pronuclei in the wild-type background (B). In the absence of paternally inherited MpBELL3 and MpBELL4, about a half of zygotes did not exhibit nuclear-enriched MpKNOX1-GFP signals at 12 HAF (D, E). At 24 HAF, weak and exclusively cytosolic GFP signals were detected in both genotypes (C, F). Bars, 10 µm. (G) A bar graph showing the ratios of zygotes with and without pronuclei-enriched MpKNOX1-GFP signals from the indicated crosses. Numbers of observed zygotes are shown above each bar.
As KNOX proteins function as transcription factors, the transient localization of MpKNOX1 in pronuclei should be critical for its role in regulating karyogamy-promoting genes. In Arabidopsis and Chlamydomonas, KNOX proteins are recruited to nuclei through interactions with BELL proteins. To examine whether paternally inherited MpBELL3 and/or MpBELL4 contribute to the pronuclear localization of MpKNOX1, we crossed gMpKNOX1-GFP females with wild-type or Mpbell3/4 males and analyzed the subcellular localization of MpKNOX1-GFP in zygotes. In contrast to zygotes derived from a cross with wild-type males, where GFP signals were preferentially detected in male and female pronuclei, approximately a half of zygotes derived from a cross with Mpbell3/4 males did not exhibit nuclear-enriched GFP signals at 12 HAF (54% and 50 % for Mpbell3/4-1ge and Mpbell3/4-2ge, respectively; Figure 6D and G). Together, these data suggest that paternally inherited MpBELL3 and/or MpBELL4 contribute to the pronuclear localization of MpKNOX1 after fertilization.
Discussion
Here, we demonstrated that MpKNOX1 is an egg-specific gene in M. polymorpha. MpKNOX1 is strongly expressed in developing and mature eggs, whereas no expression was detected in gametophytes, sperm, or sporophytes. The egg-specific expression of MpKNOX1 is in sharp contrast with the expression pattern of KNOX1 genes in another model bryophyte, P. patens, where all three KNOX1 genes are strongly expressed in sporophytes to regulate their development (Sakakibara et al., 2008). Rather, the egg-specific expression pattern of MpKNOX1 is reminiscent of that of GSM1, a KNOX gene of the unicellular green alga C. reinhardtii specifically expressed in minus gametes. Upon fertilization, GSM1 forms heterodimers with plus gamete-derived BELL protein GSP1 to activate expression of early zygote-specific genes (Joo et al., 2017; Lee et al., 2008; Nishimura et al., 2012). Accordingly, we suspected that MpKNOX1 might function as a gamete-derived zygote activator in M. polymorpha. Indeed, a majority of egg cells produced by three independent loss-of-function Mpknox1 female mutants failed to produce embryos when fertilized with wild-type sperm. By contrast, wild-type eggs fertilized with Mpknox1 sperm produced normal embryos and spores. The residual embryos produced from Mpknox1 eggs are unlikely to have formed due to paternal MpKNOX1 functions as a comparable number of embryos also arose from homozygous Mpknox1 zygotes. Rather, it is more likely that the Mpknox1 alleles generated in our study retained some functionality despite the presence of premature stop codons because a large deletion allele of Mpknox1 used by another group showed a complete penetrance when transmitted maternally (Dierschke et al., 2021). These results indicate that maternal Mpknox1 has a major, if not exclusive, contribution to embryogenesis.
The parent-of-origin effects of gene alleles can arise at several levels (Luo et al., 2014). In some cases, only one parental allele is transcribed in zygotes due to silencing of the other allele. In other cases, gene products such as proteins and small RNA molecules are synthesized in and/or carried over from gametes of a single sex. Our reporter analysis revealed that MpKNOX1 is preferentially transcribed in egg cells. By contrast, MpKNOX1-GFP accumulated in both unfertilized and fertilized eggs until 12 HAF and had mostly diminished at 24 HAF. These observations strongly argue for a mechanism in which MpKNOX1 produced in unfertilized eggs acts later in zygotes to initiate embryogenesis. This mode of action of MpKNOX1 is strikingly similar to that proposed for C. reinhardtii GSM1 (Lee et al., 2008).
How does egg-derived MpKNOX1 promote embryogenesis? We determined that in wild-type M. polymorpha karyogamy is completed only after 3–4 DAF. This slow progression of karyogamy is further delayed or arrested when maternal Mpknox1 is mutated. Observation of nuclear membrane dynamics using an MpSUN-GFP marker showed that in fertilized Mpknox1 mutant eggs karyogamy was arrested at the nuclear membrane fusion step, not the nuclear migration step. Consistent with the predicted role of MpKNOX1 as a transcription factor, MpKNOX1-GFP localized to both male and female pronuclei by 12 HAF, far before the initiation of nuclear membrane fusion. These observations suggest that gamete-derived MpKNOX1 functions in pronuclei to regulate the expression of gene(s) required for nuclear membrane fusion, thereby activating zygote development.
In both angiosperms and C. reinhardtii, the heterodimerization of KNOX and BELL is required for the nuclear translocation of these proteins, which in turn is required for them to regulate gene transcription (Hay and Tsiantis, 2010; Lee et al., 2008). Consistent with this notion, our genetic and imaging analyses suggested that sperm-derived MpBELL3/4 are required to recruit MpKNOX1 to male and female pronuclei. Our effort of examining MpBELL4 pronuclear localization, however, has been unsuccessful due to a difficulty of obtaining reporter lines, whereas a study by another group indicated interaction between MpKNOX1 and MpBELL4 in tobacco cells (Dierschke et al., 2021). Among the five BELL genes in the M. polymorpha genome, MpBELL3 and MpBELL4 are specifically expressed in antheridiophores. Mpbell3/4 mutant males could produce motile sperm, but approximately 30% of wild-type eggs fertilized with Mpbell3/4 sperm failed to produce embryos compared to ~10% of those fertilized with wild-type sperm. This partial loss of embryogenic ability correlated well with the proportion of zygotes with compromised pronuclear enrichment of MpKNOX1-GFP. These observations, together with other studies on KNOX/BELL orthologs, suggest a mechanism in which paternal MpBELL3 and/or MpBELL4 recruit maternal MpKNOX1 to pronuclei.
Interestingly, however, our genetic analyses also indicated that not only paternal but also maternal MpBELL3 and/or MpBELL4 contribute to karyogamy, even though their expression was not detected in maternal organs or egg cells. A plausible explanation for this observation is that maternally inherited MpBELL3 and/or MpBELL4 alleles become transcriptionally activated in zygotes and that MpBELL3 and/or MpBELL4 expression post-fertilization acts to replenish MpBELL proteins to ensure karyogamy with high penetrance. It should be noted, however, that 20–40% of zygotes carrying homozygous Mpbell3/4 alleles still produce embryos, and this proportion is higher than those carrying maternally inherited Mpknox1 alone (less than 10%). This lower penetrance of Mpbell3/4 may be due to redundantly acting MpBELL genes. Transcriptome data indicates weak expression of MpBELL1 and MpBELL2 in antheridiophores (Figure 5—figure supplement 1A) and preferential expression of MpBELL5 in archegonia (Figure 5—figure supplement 1B). These BELL homologs may play a minor role in promoting karyogamy.
Based on these observations, we propose a two-step model of MpBELL3/4 activity (Figure 7A). According to this model, sperm-borne MpBELL3 and MpBELL4 proteins heterodimerize with egg-derived MpKNOX1 upon fertilization and activate the transcription of zygote-specific genes as well as maternal MpBELL3 and/or MpBELL4. MpBELL3 and MpBELL4 produced de novo further activate the expression of zygote-specific genes, including genes required for karyogamy. Such feed-forward regulation would efficiently compensate for the presumably small amounts of MpBELL proteins inherited from the sperm cytosol. Considering that a recent phenotypic analysis of newly isolated gsm1 and gsp1 alleles revealed the biparental contribution of GSM1 to karyogamy in C. reinhardtii (Kariyawasam et al., 2019), our findings further emphasize the similarity of KNOX/BELL-mediated zygote activation between M. polymorpha and C. reinhardtii.
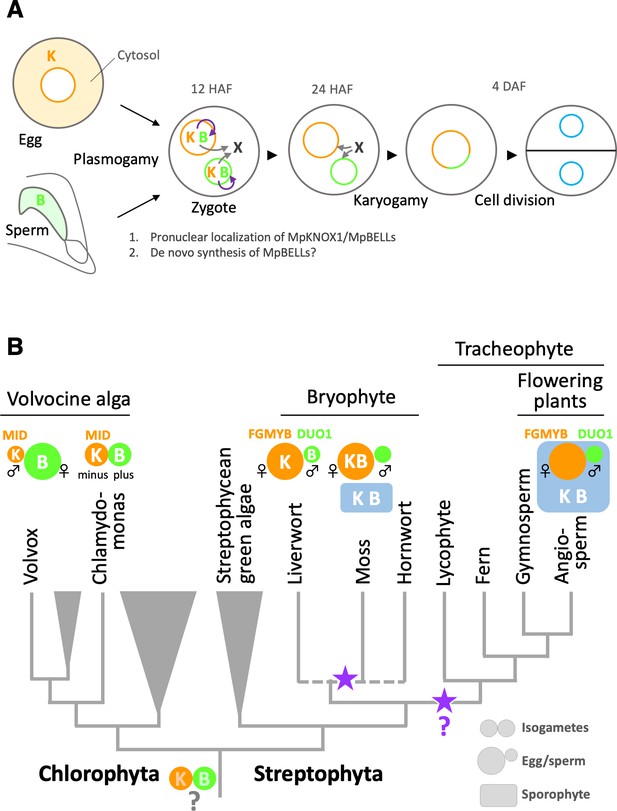
Functions and expression patterns of KNOX/BELL transcription factors in green plants.
(A) Expression patterns of KNOX and BELL proteins and their predicted role in zygote activation in M. polymorpha. ‘K’ and ‘B’ represent MpKNOX1 and MpBELL protein subunits, respectively. Orange, green, and blue circles represent female pronuclei, male pronuclei, and nuclei of embryo cells, respectively. Purple curved arrows indicate auto-amplification of BELL levels by KNOX/BELL-mediated transcriptional control. X indicates unknown karyogamy-promoting factor(s) whose expression and/or functions are activated by KNOX/BELL-mediated transcription. (B) Predicted evolutionary trajectory of KNOX/BELL expression patterns along the green plant lineages. Orange/green circles and blue rectangles represent gametes and sporophyte bodies, respectively. ‘K‘ and ‘B’ indicate the expression of KNOX and BELL proteins, respectively. Purple stars indicate the predicted positions at which the functional transition of KNOX/BELL from zygote activation to sporophyte morphogenesis occurred. Also indicated are the expression patterns of evolutionarily conserved regulators of sexual differentiation; a male-determinant factor MID of volvocine algae, and female- and male-differentiation factors, FGMYB and DUO1, respectively, of land plants. Note that the KNOX/BELL expression patterns in ancestral plants at the bottom of the tree are an inference.
While our study revealed the striking conservation of KNOX/BELL functions between M. polymorpha and C. reinhardtii, this finding is somewhat unexpected from a phylogenetic viewpoint because KNOX/BELL proteins in the model bryophyte P. patens control sporophyte development, as do KNOX/BELL proteins in angiosperms, as well as egg size (Sakakibara et al., 2008; Horst et al., 2016; Ortiz-Ramírez et al., 2017). Outside the plant kingdom, TALE TFs in yeasts and fungi promote the haploid-to-diploid transition (Goutte and Johnson, 1988; Herskowitz, 1989; Kues et al., 1992, Spit et al., 1998). Thus, perhaps the shared zygote-activating functions of KNOX/BELL in M. polymorpha and C. reinhardtii reflect an ancestral state (Bowman et al., 2016). Considering the generally accepted view of bryophyte monophyly (de Sousa et al., 2019; Puttick et al., 2018), our findings suggest that the functional transition of KNOX/BELLs from zygote activation to sporophyte morphogenesis occurred at least twice during land plant evolution, including once in the bryophyte lineage and once in the tracheophyte lineage (Figure 7B).
Our study revealed that the zygote-activating function of KNOX/BELL is conserved between C. reinhardtii and M. polymorpha, which belong to Chlorophyta and Streptophyta, respectively. These two major green plant lineages were separated more than 700 million years ago (Becker, 2013; Figure 7B). Interestingly, however, the sex-specific expression patterns of KNOX and BELL in C. reinhardtii are opposite to those in M. polymorpha. In C. reinhardtii, KNOX (GSM1) and BELL (GSP1) are expressed in isogamous minus and plus gametes (Lee et al., 2008), which directly evolved into male and female gametes, respectively, in oogamous (with small motile gametes and large immotile gametes) Volvox carteri, primarily by modifying genes acting downstream of the conserved sex-determinant protein MID (Ferris and Goodenough, 1997; Geng et al., 2014; Geng et al., 2018; Nozaki et al., 2006; Figure 7B). Results from other research groups indicate that the expression specificity of KNOX/BELL is conserved along the volvocine lineage (with minus and male gametes expressing GSM1, and plus and female gametes expressing GSP1; personal communication with Takashi Hamaji [Kyoto University, Japan] and James Umen [Donald Danforth Plant Science Center, MO]). Thus, at least in one Chlorophyta lineage, KNOX and BELL are expressed in male and female gametes, respectively, a situation opposite to that in M. polymorpha (KNOX for females and BELL for males; Figure 7B).
In land plants, oogamy likely evolved once as key regulators of gametophytic sexual differentiation, such as FGMYBs for females and DUO POLLEN 1 (DUO1) for males, are shared between M. polymorpha and Arabidopsis (see Hisanaga et al., 2019b for a review; Figure 7B). Theoretical analyses predicted that anisogamy evolved through disruptive selection, where an increased volume of one type of gamete favors zygote fitness, allowing the other gamete type to increase in number at the expense of volume (Parker et al., 1972). Thus, in ancestral green plants, KNOX/BELL expression specificity did not affect gamete morphology or function. This notion is consistent with our observation that neither MpKNOX1 nor MpBELL3/4 contribute to gamete development or function in extant M. polymorpha.
In summary, our study revealed a critical role of KNOX and BELL in zygote activation in the model bryophyte M. polymorpha. This is in stark contrast with a well-recognized role of KNOX/BELL in sporophytic meristem maintenance in angiosperms and another model bryophyte P. patens. Rather, striking conservation of KNOX/BELL functions in the promotion of karyogamy across phylogenetically distant M. polymorpha and C. reinhardtii suggests that functions of KNOX/BELL heterodimers shifted from zygote activation to sporophyte development as land plants evolved. This view is consistent with a proposed evolutionary scenario of HD proteins in broader eukaryotic taxa, including fungi and metazoans (Bowman et al., 2016).
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Marchantia polymorpha) | MpKNOX1 | MalpolBase | Mp5g01600 | |
| Gene (Marchantia polymorpha) | MpBELL1 | MalpolBase | Mp8g18310 | |
| Gene (Marchantia polymorpha) | MpBELL2 | MalpolBase | Mp4g09650 | |
| Gene (Marchantia polymorpha) | MpBELL3 | MalpolBase | Mp8g02970 | |
| Gene (Marchantia polymorpha) | MpBELL4 | MalpolBase | Mp8g07680 | |
| Gene (Marchantia polymorpha) | MpBELL5 | MalpolBase | Mp5g11060 | |
| Gene (Marchantia polymorpha) | MpSUN | MalpolBase | Mp5g02400 | |
| Gene (Marchantia polymorpha) | MpECpro | MalpolBase | Mp5g18000 | |
| Strain, strain background (Marchantia polymorpha, male) | Tak-1 | DOI:10.1016/j.cell.2017.09.030 | Male wild-type strain of Marchantia | |
| Strain, strain background (Marchantia polymorpha, female) | Tak-2 | DOI:10.1016/j.cell.2017.09.030 | Female wild-type strain of Marchantia | |
| Recombinant DNA reagent | pMpGE_En03 | Addgene | RRID:Addgene_71535 | Entry plasmid for gRNA |
| Recombinant DNA reagent | pMpGE010 | Addgene | RRID:Addgene_71536 | Destination plasmid for CRISPR/Cas9 |
| Recombinant DNA reagent | pMpGE_En04 | DOI:10.15252/embj.2018100240 | Entry plasmid for gRNA | |
| Recombinant DNA reagent | pBC-GE12 | DOI:10.15252/embj.2018100240 | Entry plasmid for gRNA | |
| Recombinant DNA reagent | pBC-GE23 | DOI:10.15252/embj.2018100240 | Entry plasmid for gRNA | |
| Recombinant DNA reagent | pBC-GE34 | DOI:10.15252/embj.2018100240 | Entry plasmid for gRNA | |
| Recombinant DNA reagent | pMpGE017 | DOI:10.15252/embj.2018100240 | Destination plasmid for Cas9 nickase | |
| Recombinant DNA reagent | pMpGE010_MpKNOX1ge | This paper | Plasmid to create Mpknox1 mutants | |
| Recombinant DNA reagent | pMpGE017_MpBELL4ge-MpBELL3ge | This paper | Plasmid to create Mpbell3/4 mutants | |
| Recombinant DNA reagent | pMpSL30 | DOI:10.15252/embj.2018100240 | Destination plasmid | |
| Recombinant DNA reagent | pMpSL30_MpKNOX1pro-H2B-GFP-3’MpKNOX1 | This paper | MpKNOX1 promoter reporter | |
| Recombinant DNA reagent | pMpSL30_gMpKNOX1-GFP | This paper | MpKNOX1 complementation | |
| Recombinant DNA reagent | pMpSL30_ ECpro-MpSUN-GFP | This paper | Egg cell-specific nuclear envelop marker | |
| Genetic reagent (Marchantia polymorpha) | Mpknox1-1ge | This paper | Mpknox1 CRISPR mutant obtained from sporeling transformation | |
| Genetic reagent (Marchantia polymorpha) | Mpknox1-2ge | This paper | Mpknox1 CRISPR mutant obtained from sporeling transformation | |
| Genetic reagent (Marchantia polymorpha) | Mpknox1-3ge | This paper | Mpknox1 CRISPR mutant obtained from sporeling transformation | |
| Genetic reagent (Marchantia polymorpha) | Mpbell3/4-1ge | This paper | Mpbell3/4 CRISPR mutant obtained from sporeling transformation | |
| Genetic reagent (Marchantia polymorpha) | Mpbell3/4-2ge | This paper | Mpbell3/4 CRISPR mutant obtained from sporeling transformation | |
| Genetic reagent (Marchantia polymorpha) | MpKNOX1pro:H2B-GFP/WT ♀ | This paper | pMpSL30_MpKNOX1pro-H2B-GFP-3′MpKNOX1 was transformed into Tak-2 | |
| Genetic reagent (Marchantia polymorpha) | gMpKNOX1-GFP/Mpknox1-1ge♀ | This paper | pMpSL30_gMpKNOX1-GFP was transformed into Mpknox1-1ge | |
| Genetic reagent (Marchantia polymorpha) | ECpro:MpSUN-GFP | This paper | pMpSL30_ ECpro-MpSUN-GFP was transformed into Tak-2 or Mpknox1-2ge | |
| Genetic reagent (Marchantia polymorpha) | Mprkd-1 | DOI:10.1016/j.cub.2016.05.013 | ||
| Genetic reagent (Marchantia polymorpha) | Mprkd-3 | DOI:10.1016/j.cub.2016.05.013 | ||
| Commercial assay or kit | RNeasy Plant Mini Kit | Qiagen | 74904 | |
| Commercial assay or kit | TruSeq RNA Sample Prep Kit v2 | Illumina | RS-122-2001 | |
| Commercial assay or kit | Gateway LR Clonase II Enzyme mix | Thermo Fisher Scientific | 11791020 | |
| Software, algorithm | TopHat ver. 2.0.14 | doi:10.1093/bioinformatics/btp120 | RRID:SCR_013035 | |
| Software, algorithm | DESeq2 | doi:10.1186/s13059-014-0550-8 | RRID:SCR_015687 | |
| Software, algorithm | TCC | DOI:10.1186/1471-2105-14-219 | RRID:SCR_001779 |
Plant materials
Request a detailed protocolMarchantia polymorpha L. subsp. ruderalis accessions Takaragaike 1 (Tak-1) and Takaragaike 2 (Tak-2; Ishizaki et al., 2016) were used as the wild-type male and female, respectively. Plants were cultured on half-strength Gamborg’s B5 medium solidified with 1% (w/v) agar under continuous white light at 22°C. To induce reproductive development, 10-day-old thalli were transferred to a pot containing vermiculite and grown under white light supplemented with far-red illumination generated by LED (VBL-TFL600-IR730; IPROS Co., Tokyo, Japan).
Archegonium sampling, RNA extraction, and Illumina sequencing
Request a detailed protocolMature archegonia were manually dissected from wild-type archegoniophores and divided into four pools, each composed of approximately 1000 archegonia. Mature archegonia of Mprkd-1 and Mprkd-3 (Koi et al., 2016) were collected and each divided into two pools of approximately 1000 archegonia. Total RNA was extracted from the samples with an RNeasy Plant Mini Kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s protocol. The quality and quantity of the RNA were evaluated using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA) and a RNA6000 Nano Kit (Agilent Technologies, Santa Clara, CA). Sequence libraries were constructed with a TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA) according to the manufacturer’s protocol. The quality of each library was examined using a Bioanalyzer with High Sensitivity DNA Kit (Agilent Technologies) and a KAPA Library Quantification Kit for Illumina (Roche Diagnostics, Basel, Switzerland). An equal amount of each library was mixed to generate a 2 nM pooled library. Next-generation sequencing was performed using the HiSeq 1500 platform (Illumina) to generate 126-nt single-end data. Sequence data have been deposited at the DDBJ BioProject and BioSample databases under accession numbers PRJDB9329 and SAMD00205647-SAMD00205654, respectively.
Data analysis
Request a detailed protocolRead data were mapped to the genome sequence of M. polymorpha v3.1 using TopHat ver. 2.0.14 (Trapnell et al., 2009) with default parameters. FPKM values were calculated using the DESeq2 package in R (Love et al., 2014). Differentially expressed genes (DEGs) were identified using the TCC package in R (Sun et al., 2013) with a criterion of false discovery rate (FDR) < 0.01. Candidate egg-specific genes were identified by filtering the DEGs with a threshold of Mprkd/WT ratio <−3 and FPKM in WT > 1.
Semi in vitro culture and genetic crossing
Request a detailed protocolMature archegoniophores and antheridiophores were separated from thalli and collected into a 5 mL plastic tube containing 3 mL of water. Following co-culture for 1 hr under white light at 22°C, the archegoniophores were transferred to new 5 mL plastic tubes containing 3 mL of water and cultured under white light at 22°C prior to observation.
Microscopy
Request a detailed protocolTo observe MpSUN-GFP expression, archegonia were excised under a dissecting microscope and mounted in half-strength Gamborg’s B5 liquid medium. To observe pronuclei, archegoniophores were dissected and soaked in PFA fixative solution (4% [w/v] paraformaldehyde, 1 µg/mL DAPI, and 0.01% [v/v] Triton X-100 in 1× PBS buffer). The samples were briefly vacuum-infiltrated four times and incubated for 1 hr at room temperature with gentle shaking. The samples were washed twice with 1× PBS and cleared by incubating in ClearSee solution containing 1 µg/mL DAPI for 2–3 days (Kurihara et al., 2015). The cleared samples were observed under a Nikon C2 confocal laser-scanning microscope (Nikon Instech, Tokyo, Japan). Sperm cells were stained with DAPI as described previously (Hisanaga et al., 2019a).
RT-PCR
Request a detailed protocolRNA extraction, cDNA synthesis, and RT-PCR were performed as described previously (Hisanaga et al., 2019a) using the primer sets listed in Supplementary file 2.
DNA constructs
The plasmids used in this study were constructed using the Gateway cloning system (Ishizaki et al., 2015), the SLiCE method (Motohashi, 2015), or Gibson assembly (Gibson et al., 2009). The primers used for DNA construction are listed in Supplementary file 2.
pMpGE010_MpKNOX1ge
Request a detailed protocolA DNA fragment producing MpKNOX1-tageting gRNAs was prepared by annealing a pair of synthetic oligonucleotides (MpKNOX1ge01Fw/MpKNOX1ge01Rv). The fragment was inserted into the BsaI site of pMpGE_En03 (cat. no. 71535, Addgene, Cambridge, MA) to yield pMpGE_En03-MpKNOX1ge, which was transferred into pMpGE010 (cat. no. 71536, Addgene) (Sugano et al., 2018) using the Gateway LR reaction (Thermo Fisher Scientific, Waltham, MA) to generate pMpGE010_MpKNOX1ge.
pMpGE017_MpBELL4ge-MpBELL3ge
Request a detailed protocolTo construct a plasmid to disrupt MpBELL3 and MpBELL4 simultaneously, four oligonucleotide pairs (MpBELL4-ge1-Fw/MpBELL4-ge1-Rv, MpBELL4-ge2-Fw/MpBELL4-ge2-Rv, MpBELL3-ge3-Fw/MpBELL3-ge3-Rv, and MpBELL3-ge4-Fw/MpBELL3-ge4-Rv) were annealed and cloned into pMpGE_En04, pBC-GE12, pBC-GE23, and pBC-GE34 to yield pMpGE_En04-MpBELL4-ge1, pBC-GE12-MpBELL4-ge-2, pBC-GE23-MpBELL3-ge3, and pBC-GE34-MpBELL3-ge4, respectively. These four plasmids were assembled via BglI restriction sites and ligated to yield pMpGE_En04-MpBELL4-ge12-MpBELL3-ge34. The resulting DNA fragment containing four MpU6promoter-gRNA cassettes was transferred into pMpGE017 using the Gateway LR reaction to yield pMpGE017_MpBELL4-ge12-MpBELL3-ge34.
pMpSL30_MpKNOX1pro-H2B-GFP-3′MpKNOX1
Request a detailed protocolA 6.3 kb genomic fragment spanning the 5.3 kb 5′ upstream sequence plus the 1 kb 5′-UTR of MpKNOX1 was amplified from Tak-2 genomic DNA using the primers H-MpKNOX1pro-Fw and SmaI-MpKNOX1pro-Rv. A vector backbone containing the GFP-coding sequence was prepared by digesting pAN19_SphI-35S-lox-IN-lox-NosT-SmaI-GFP-NaeI (a kind gift from Dr. Shunsuke Miyashima) with SphI and SmaI. The two fragments were assembled using the SLiCE reaction to yield pAN19_MpKNOX1pro-SmaI-GFP-NaeI. The histone H2B-coding sequence from Arabidopsis was amplified from the pBIN41_DUO1pro-H2B-YFP-nos vector (Hisanaga, unpublished) using the primers KNOXp-H2B-Fw and GFP-H2B-Rv. The fragment was inserted into the SmaI site of pAN19_MpKNOX1pro-SmaI-GFP-NaeI by the SLiCE reaction, yielding pAN19_MpKNOX1pro-H2B-GFP-NaeI. A 4 kb fragment containing the 0.5 kb 3′-UTR plus 3.5 kb 3′-flanking sequences of MpKNOX1 was amplified from Tak-2 genomic DNA using the primers G-MpKNOX1ter-Fw and E-MpKNOX1ter-Rv. The fragment was inserted into the NaeI site of pAN19_MpKNOX1pro-H2B-GFP-NaeI by the SLiCE reaction, yielding pAN19_MpKNOX1pro-H2B-GFP-3’MpKNOX1. The MpKNOX1pro-H2B-GFP-3’MpKNOX1 fragment was excised from pAN19_MpKNOX1pro-H2B-GFP-3’MpKNOX1 by digestion with AscI and inserted into pMpSL30 (Hisanaga et al., 2019a) to yield pMpSL30_MpKNOX1pro-H2B-GFP-3′MpKNOX1.
gMpKNOX1-GFP
Request a detailed protocolA 3.4 kb genomic fragment spanning the entire exon and intron region of MpKNOX1 was amplified from Tak-2 genomic DNA using the primers gMpKNOX1-Fw and gMpKNOX1-Rv. The fragment was inserted into the SmaI site of pAN19_MpKNOX1pro-SmaI-GFP-NaeI by the SLiCE reaction to yield pAN19_MpKNOX1pro-MpKNOX1-GFP-NaeI. A 4 kb fragment containing the 0.5 kb 3′-UTR and 3.5 kb 3′-flanking sequences of MpKNOX1 was amplified from Tak-2 genomic DNA using the primers G-MpKNOX1ter-Fw and E-MpKNOX1ter-Rv. The fragment was inserted into the NaeI site of pAN19_MpKNOX1pro-MpKNOX1-GFP-NaeI by the SLiCE reaction, yielding pAN19_gMpKNOX1-GFP. The gMpKNOX1-GFP fragment was excised from pAN19_gMpKNOX1-GFP by digestion with AscI and inserted into pMpSL30 to yield pMpSL30_gMpKNOX1-GFP.
ECpro:MpSUN-GFP
Request a detailed protocolA 5 kb genomic fragment spanning the 3.4 kb 5′ upstream and 1.6 kb 5′-UTR sequences of Mp5g18000 was amplified from Tak-2 genomic DNA using the primers H-ECpro-Fw and G-SpeI-ECpro-Rv. The fragment was inserted into the SpeI site of pAN19_SpeI-GFP-NaeI by the SLiCE reaction, yielding pAN19_ECpro-SpeI-GFP-NaeI. The 1.5 kb MpSUN (Mp5g02400)-coding sequence was amplified from a cDNA library of M. polymorpha using the primers E-MpSUN-Fw and G-MpSUN-Rv. The fragment was inserted into the SpeI site of pAN19_ECpro-SpeI-GFP-NaeI by the SLiCE reaction, yielding pAN19_ECpro-MpSUN-GFP. The ECpro-MpSUN-GFP fragment was excised from pAN19_ECpro-MpSUN-GFP by digestion with AscI and inserted into pMpSL30 to yield pMpSL30_ ECpro-MpSUN-GFP.
Generation of transgenic M. polymorpha
Request a detailed protocolGenome editing constructs were introduced into M. polymorpha sporelings as described previously (Ishizaki et al., 2008). Other constructs were introduced into regenerating thalli (Kubota et al., 2013) or gemmae using the G-AgarTrap method (Tsuboyama et al., 2018).
Data availability
Sequence data have been deposited at the DDBJ BioProject and BioSample databases under accession numbers PRJDB9329 and SAMD00205647-SAMD00205654, respectively.
-
EBI European Nucleotide ArchiveID PRJDB9329. Transcriptome analysis of archegonia in Marchantia wild type and Mprkd mutant.
-
NCBI BioProjectID PRJNA251267. Marchantia polymorpha strain:Takaragaike-1 (male) Takaragaike-1 (female, Takaragaike-2 was back-crossed four times to Takaragaike-1) (liverwort).
References
-
Snow ball earth and the split of Streptophyta and ChlorophytaTrends in Plant Science 18:180–183.https://doi.org/10.1016/j.tplants.2012.09.010
-
A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motifThe Journal of Biological Chemistry 270:31178–31188.https://doi.org/10.1074/jbc.270.52.31178
-
Evolution in the cycles of lifeAnnual Review of Genetics 50:133–154.https://doi.org/10.1146/annurev-genet-120215-035227
-
BookEvolution and co-option of developmental regulatory networks in early land plantsIn: Grossniklaus U, editors. Current Topics in Developmental Biology, Vol. 31. London: Academic Press. pp. 35–53.https://doi.org/10.1016/bs.ctdb.2018.10.001
-
Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotesEvolution & Development 9:212–219.https://doi.org/10.1111/j.1525-142X.2007.00153.x
-
The Development of the Sexual Organs and Sporogonium of Marchantia polymorphaBulletin of the Torrey Botanical Club 35:321.https://doi.org/10.2307/2485335
-
Gamete nuclear migration in animals and plantsFrontiers in Plant Science 10:517.https://doi.org/10.3389/fpls.2019.00517
-
Nonreciprocal complementation of KNOX gene function in land plantsThe New Phytologist 216:591–604.https://doi.org/10.1111/nph.14318
-
Transcriptional framework of male gametogenesis in the liverwort Marchantia polymorpha LPlant & Cell Physiology 57:325–338.https://doi.org/10.1093/pcp/pcw005
-
Molecular genetic tools and techniques for Marchantia polymorpha ResearchPlant & Cell Physiology 57:262–270.https://doi.org/10.1093/pcp/pcv097
-
Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalliBioscience, Biotechnology, and Biochemistry 77:167–172.https://doi.org/10.1271/bbb.120700
-
The expression and roles of parent-of-origin genes in early embryogenesis of angiospermsFrontiers in Plant Science 5:729.https://doi.org/10.3389/fpls.2014.00729
-
A comprehensive classification and evolutionary analysis of plant homeobox genesMolecular Biology and Evolution 26:2775–2794.https://doi.org/10.1093/molbev/msp201
-
Males evolved from the dominant isogametic mating typeCurrent Biology 16:R1018–R1020.https://doi.org/10.1016/j.cub.2006.11.019
-
The origin and evolution of gamete dimorphism and the male-female phenomenonJournal of Theoretical Biology 36:529–553.https://doi.org/10.1016/0022-5193(72)90007-0
-
Marchantia polymorpha: Taxonomy, phylogeny and morphology of a model systemPlant & Cell Physiology 57:230–256.https://doi.org/10.1093/pcp/pcv192
-
Tophat: Discovering splice junctions with RNA-seqBioinformatics 25:1105–1111.https://doi.org/10.1093/bioinformatics/btp120
-
SUN-domain proteins: “Velcro” that links the nucleoskeleton to the cytoskeletonNature Reviews. Molecular Cell Biology 7:782–788.https://doi.org/10.1038/nrm2003
-
The fine structure of oogenesis in Marchantia polymorphaAmerican Journal of Botany 61:499–512.https://doi.org/10.1002/j.1537-2197.1974.tb10789.x
Article and author information
Author details
Funding
Japan Society for the Promotion of Science (KAKENHI 17J08430)
- Tetsuya Hisanaga
Ministry of Education, Culture, Sports, Science and Technology (KAKENHI 25113007)
- Keiji Nakajima
Japan Society for the Promotion of Science (KAKENHI 18K06285)
- Shohei Yamaoka
Ministry of Education, Culture, Sports, Science and Technology (KAKENHI 17H05841)
- Shohei Yamaoka
Ministry of Education, Culture, Sports, Science and Technology (KAKENHI 25113009)
- Takayuki Kohchi
Japan Society for the Promotion of Science (KAKENHI 17H07424)
- Takayuki Kohchi
European Commission (Marie Sklodowska-Curie grant agreement 847548)
- Tetsuya Hisanaga
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank Masako Kanda for technical assistance and Shunsuke Miyashima for DNA materials. This work was supported by MEXT KAKENHI grants 17J08430 to TH, 25113007 to KN, 17H05841 and 18K06285 to SY, and 25113009 and 17H07424 to TK. TH was supported by a JSPS Fellowship for Young Scientists and a funding from the European Union’s Framework Programme for Research and Innovation Horizon 2020 (2014–2020) under the Marie Curie Skłodowska Grant Agreement Nr. 847548.
Version history
- Received: March 20, 2020
- Preprint posted: April 9, 2020 (view preprint)
- Accepted: September 2, 2021
- Version of Record published: September 28, 2021 (version 1)
- Version of Record updated: December 15, 2021 (version 2)
Copyright
© 2021, Hisanaga et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,696
- views
-
- 345
- downloads
-
- 19
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Developmental Biology
- Evolutionary Biology
Despite rapid evolution across eutherian mammals, the X-linked MIR-506 family miRNAs are located in a region flanked by two highly conserved protein-coding genes (SLITRK2 and FMR1) on the X chromosome. Intriguingly, these miRNAs are predominantly expressed in the testis, suggesting a potential role in spermatogenesis and male fertility. Here, we report that the X-linked MIR-506 family miRNAs were derived from the MER91C DNA transposons. Selective inactivation of individual miRNAs or clusters caused no discernible defects, but simultaneous ablation of five clusters containing 19 members of the MIR-506 family led to reduced male fertility in mice. Despite normal sperm counts, motility, and morphology, the KO sperm were less competitive than wild-type sperm when subjected to a polyandrous mating scheme. Transcriptomic and bioinformatic analyses revealed that these X-linked MIR-506 family miRNAs, in addition to targeting a set of conserved genes, have more targets that are critical for spermatogenesis and embryonic development during evolution. Our data suggest that the MIR-506 family miRNAs function to enhance sperm competitiveness and reproductive fitness of the male by finetuning gene expression during spermatogenesis.






