Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases
Figures
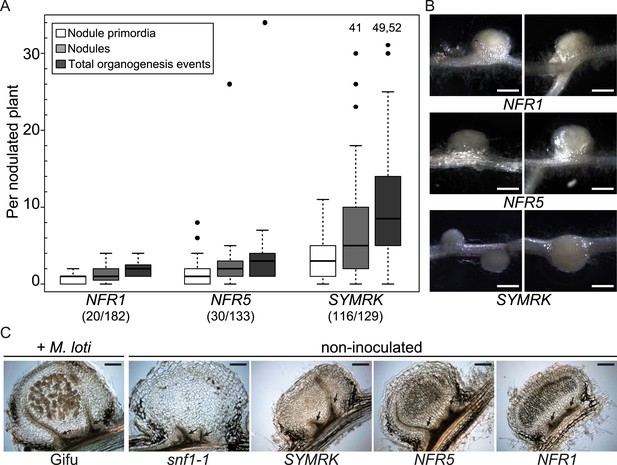
Symbiotic RLKs mediate spontaneous formation of root nodules.
Hairy roots of L. japonicus Gifu wild-type transformed with the empty vector (EV), pUB:NFR1-mOrange (NFR1), pUB:NFR5-mOrange (NFR5), or pUB:SYMRK-mOrange (SYMRK) were generated. (A) Plot represents the numbers of nodule primordia (white), nodules (light grey) and total organogenesis events (dark grey; nodules and nodule primordia) per nodulated plant formed in the absence of rhizobia at 60 dpt. Number of nodulated plants per total plants is specified under each line label. Black dots, data points outside 1.5 interquartile range (IQR) of the upper quartile; numbers above upper whiskers indicate the values of individual data points outside of the plotting area. Bold black line, median; box, IQR; whiskers, lowest/highest data point within 1.5 IQR of the lower/upper quartile. Plants transformed with the empty vector did not develop spontaneous nodules. (B) Pictures of spontaneous nodules on hairy roots expressing the indicated transgenes taken 60 dpt. Bars, 1 mm. (C) Micrographs of sections of spontaneous nodules on hairy roots expressing the indicated transgenes harvested at 60 dpt. Spontaneous nodules of 10-week-old snf1-1 mutant plants were used as controls. Nodules of 10-week-old untransformed L. japonicus wild-type Gifu 6 weeks after inoculation with M. loti MAFF303099 DsRED contained cortical cells filled with bacteria (brown colour) that are absent in spontaneous nodules. Arrows point to peripheral vascular bundles. Longitudinal 40 mm sections. Bars, 150 µm.
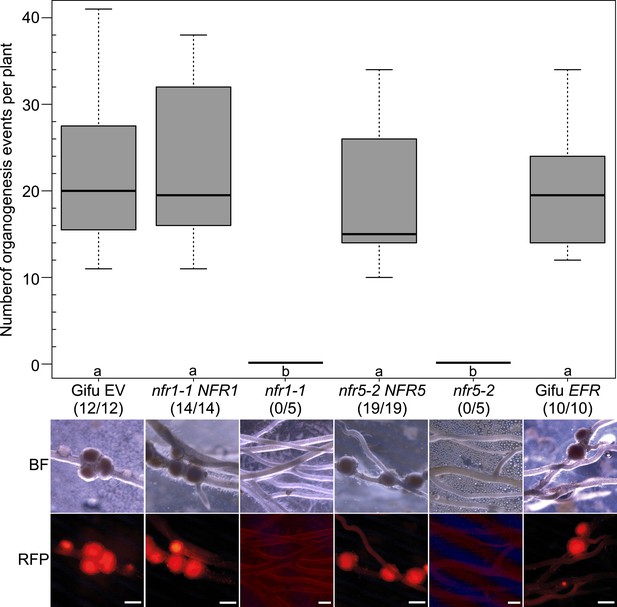
Expression of NFR1 and NFR5 from the Ubiquitin promoter restores nodulation in the nfr1-1 and nfr5-2 mutants, respectively.
Hairy roots of L. japonicus Gifu wild-type transformed with the empty vector (EV) or with pUB:EFR-mOrange (EFR), the nfr1-1 mutant transformed with pUB:NFR1-mOrange (NFR1) or the nfr5-2 mutant transformed with pUB:NFR5-mOrange (NFR5) were generated. Untransformed nfr1-1 and nfr5-2 mutant plants served as control. Plot represents the number of organogenesis events (nodules and nodule primordia) per plant formed 15 days post inoculation with M. loti DsRED. Numbers below each line label indicate the number of nodulated plants per total analysed plants. Representative pictures are shown. BF, bright field; RFP, RFP filter. Bars, 1 mm. Bold black line, median; box, IQR; whiskers, lowest/highest data point within 1.5 IQR of the lower/upper quartile. A Kruskal–Wallis test followed by false discovery rate correction was performed. Different letters indicate significant differences. p < 0.05.
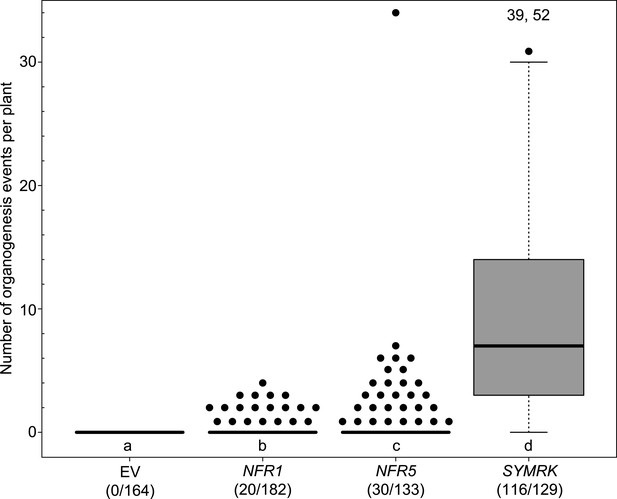
Statistical analysis of spontaneous root nodule formation.
Hairy roots of L japonicus Gifu wild-type transformed with the empty vector (EV), pUB:NFR1-mOrange (NFR1), pUB:NFR5-mOrange (NFR5), or pUB:SYMRK-mOrange (SYMRK) were generated. Plot represents the numbers of organogenesis events (nodules and nodule primordia) per plant formed in the absence of rhizobia at 60 dpt. Number of nodulated plants per total plants is specified under each line label. Black dots, data points outside 1.5 IQR of the upper quartile; numbers above upper whiskers indicate the values of individual data points outside of the plotting area. Bold black line, median; box, IQR; whiskers, lowest/highest data point within 1.5 IQR of the lower/upper quartile. A Kruskal–Wallis test followed by false discovery rate correction was performed. Different letters indicate significant differences. p < 0.05.
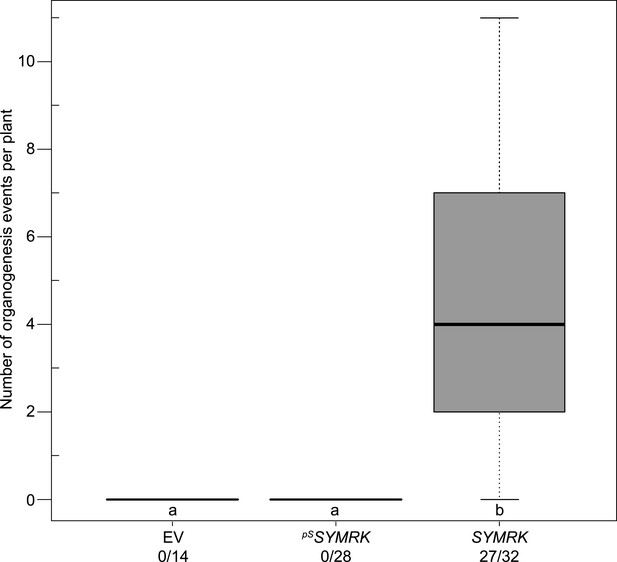
Expression of SYMRK from the native SYMRK promoter does not mediate spontaneous formation of root nodules.
Hairy roots of L. japonicus symrk-3 transformed with the empty vector (EV), pUB:SYMRK-mOrange (SYMRK), or pSYMRK:SYMRK-RFP ( pS SYMRK) were generated. Plot represents the number of total organogenesis events (nodules and nodule primordia) per plant formed in the absence of rhizobia at 21 dpt. Number of nodulated plants per total plants is specified under each line label. Bold black line, median; box, IQR; whiskers, lowest/highest data point within 1.5 IQR of the lower/upper quartile. A Kruskal–Wallis test followed by false discovery rate correction was performed. Different letters indicate significant differences. p < 0.05.
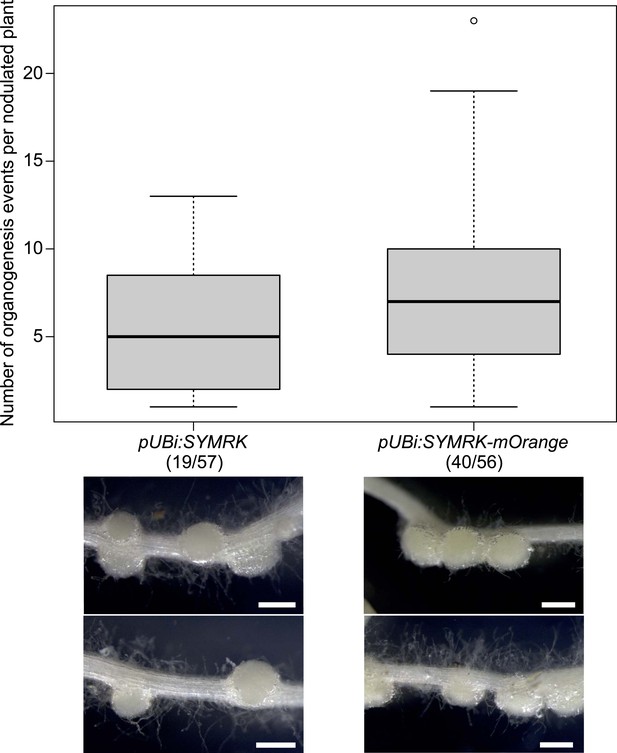
Expression of non-tagged SYMRK from the Ubiquitin promoter induces spontaneous formation of root nodules.
Hairy roots of L. japonicus symrk-3 transformed with pUBi:SYMRK (untagged) or pUBi:SYMRK-mOrange (C-terminally tagged) were generated. Plot represents the number of total organogenesis events (nodules and primordia) per nodulated plant formed in the absence of rhizobia at 42 dpt. Number of nodulated plants per total plants is specified under each line label. Dot, data point outside 1.5 interquartile range of the upper quartile. Bold black line, median; box, IQR; whiskers, lowest/highest data point within 1.5 IQR of the lower/upper quartile. Plants non-transformed or transformed with the empty vector did not develop spontaneous nodules. A Kruskal–Wallis test followed by false discovery rate correction was performed for total organogenesis events per nodulated root system (p-value of 0.16) and for total organogenesis events per transformed root system (p-value of 1.2e-05). Numbers below each line label indicate the number of nodulated plants per total analysed plants. Representative pictures are shown. Bars, 0.5 mm.
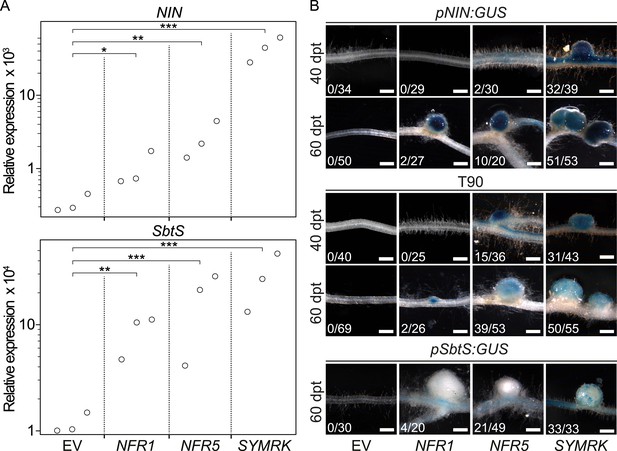
Symbiotic RLKs mediate spontaneous symbiosis-related signal transduction.
Hairy roots of L. japonicus Gifu wild-type (A) or of three stable transgenic L. japonicus Gifu reporter lines (B)—carrying either the T90 reporter fusion, a NIN promoter:GUS fusion (pNIN:GUS), or a SbtS promoter:GUS fusion (pSbtS:GUS)—transformed with the empty vector (EV), pUB:NFR1-mOrange (NFR1), pUB:NFR5-mOrange (NFR5), or pUB:SYMRK-mOrange (SYMRK) were generated. (A) Relative expression of NIN or SbtS at 40 dpt was determined in three biological replicates for each treatment via qRT-PCR. Transcript levels in each replicate were determined through technical duplicates. Expression was normalized with the house keeping genes EF1alpha and Ubiquitin. Circles indicate expression relative to the EF1alpha gene. A Dunnett's test was performed comparing the transcript levels of NIN or SbtS detected for each treatment with those detected in the empty vector samples. Stars indicate significant differences from the EV control. *, p < 0.05; **, p < 0.01; ***, p < 0.001. (B) β-glucuronidase (GUS) activity was analysed by histochemical staining with 5‐bromo‐4‐chloro‐3‐indolyl glucuronide (X-Gluc) 40 and 60 dpt. Representative root sections are shown. Number of plants with detectable GUS activity per number of total plants is indicated. Bars, 500 μm.
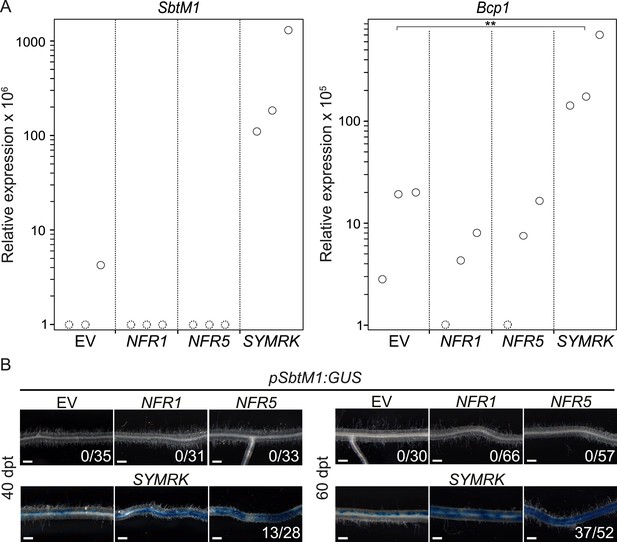
SYMRK mediates spontaneous AM-related signal transduction.
Hairy roots of L. japonicus Gifu wild-type (A) or a stable transgenic L. japonicus MG20 reporter line carrying a SbtM1 promoter:GUS fusion (pSbtM1:GUS) (B) transformed with the empty vector (EV), pUB:NFR1-mOrange (NFR1), pUB:NFR5-mOrange (NFR5), or pUB:SYMRK-mOrange (SYMRK) were generated. (A) Relative expression of SbtM1 or Bcp1 at 40 dpt was determined in three biological replicates for each treatment via qRT-PCR. Transcript levels in each replicate were determined through technical duplicates. Expression was normalized with the house keeping genes EF1alpha and Ubiquitin. Circles indicate expression relative to the EF1alpha gene. Dashed circles indicate that no transcripts could be detected for this sample. Samples in which the indicated transcript could not be detected were floored to 1. A Dunnett's test was performed comparing the transcript levels of Bcp1 detected for each treatment with those detected in the empty vector samples. Stars indicate significant differences. **, p < 0.01. (B) GUS activity was analysed by histochemical staining with X-Gluc 40 and 60 dpt. Representative root sections are shown. Number of plants with detectable GUS activity per total plants is indicated. Bars, 500 μm.
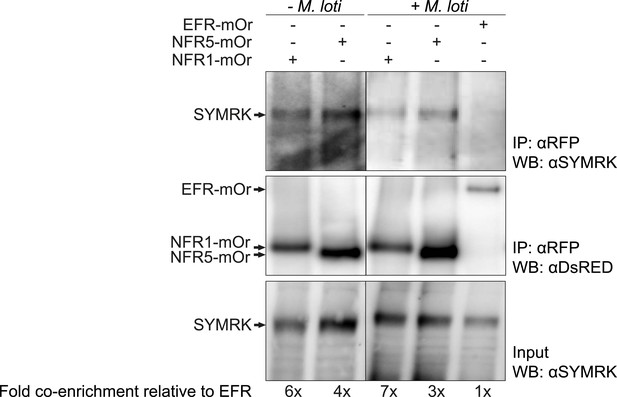
SYMRK associates with NFR1 and NFR5 in Lotus japonicus roots.
Hairy roots of L. japonicus Gifu wild-type roots expressing NFR1-mOrange (NFR1-mOr), NFR5-mOrange (NFR5-mOr), or EFR-mOrange (EFR-mOr) under the control of the Ubiquitin promoter were extracted 10 days post inoculation with M. loti DsRED or mock treatment. mOrange fusions were affinity bound with RFP magneto trap, and immuno-enrichment was monitored by immunoblot with and antiDsRED antibody. Co-enrichment of endogenous SYMRK protein was monitored by immunoblot with an antiSYMRK antibody. Numbers below the western blot panels indicate the fold co-enrichment of SYMRK by NFR1 or NFR5 relative to the amount of SYMRK co-enriched with EFR. mOr, mOrange; IE, immuno-enrichment; WB, western blot.
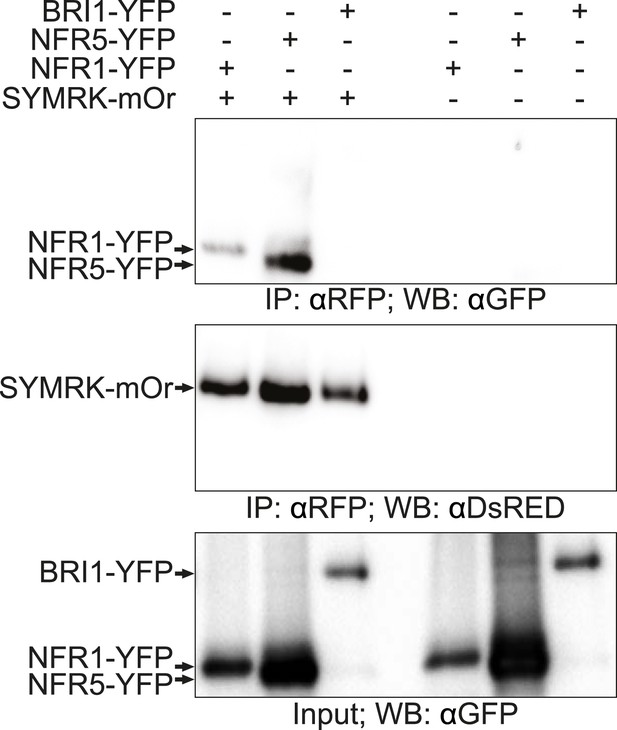
Full-length SYMRK associates with NFR1 and NFR5 in Nicotiana benthamiana leaves.
N. benthamiana leaves were transiently co-transformed with constructs expressing NFR1-YFP, NFR5-YFP, or BRI1-YFP together with SYMRK-mOrange under the control of the CaMV 35S promoter. Leaf discs expressing the respective constructs were extracted 3 dpt. SYMRK-mOrange was immuno-enriched with RFP magnetotrap and monitored by immunoblot with an antiDsRED antibody. Co-enrichment of NFR1-YFP, NFR5-YFP, or BRI1-YFP was monitored by immunoblot with an antiGFP antibody. mOr, mOrange; IE, immuno-enrichment; WB, western blot.
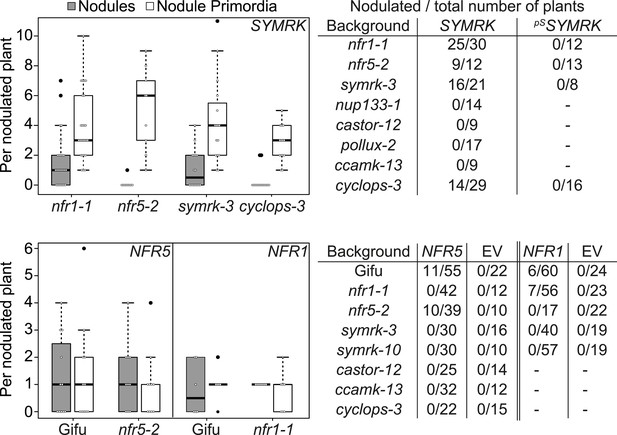
Epistatic relationships between symbiotic RLK genes and common symbiosis genes.
Hairy roots of L. japonicus Gifu wild-type and different symbiosis defective mutants transformed with pUB:SYMRK-mOrange (SYMRK) or pSYMRK:SYMRK-RFP ( pS SYMRK) (upper panel), or the empty vector (EV), pUB:NFR1-mOrange (NFR1) or pUB:NFR5-mOrange (NFR5) (lower panel) were generated. Plots represent the numbers of nodules (grey) and nodule primordia (white) per nodulated plant formed in the absence of rhizobia at 40 (SYMRK) and 60 (NFR5 + NFR1) dpt. White circles indicate individual organogenesis events. Black dots, data points outside 1.5 IQR of the upper/lower quartile; bold black line, median; box, IQR; whiskers, lowest/highest data point within 1.5 IQR of the lower/upper quartile. Table, fraction of nodulated per total number of plants. Plants transformed with pSYMRK:SYMRK-RFP or the empty pUB vector did not develop spontaneous nodules.
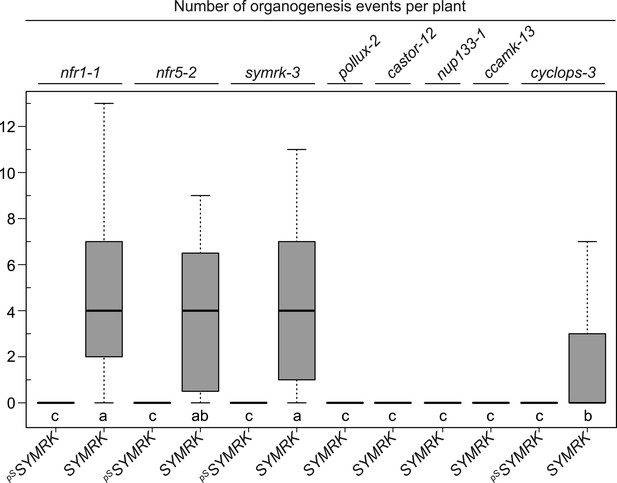
SYMRK-mediated spontaneous organogenesis events in nfr1-1, nfr5-2, and common symbiosis mutants.
Hairy roots of different symbiosis defective mutants transformed with pUB:SYMRK-mOrange (SYMRK) or pSYMRK:SYMRK-RFP ( pS SYMRK) were generated. Plot represents the numbers of organogenesis events (nodules and nodule primordia) per plant formed in the absence of rhizobia at 40 dpt. Bold black line, median; box, IQR; whiskers, lowest/highest data point within 1.5 IQR of the lower/upper quartile. A Kruskal–Wallis test followed by false discovery rate correction was performed. Different letters indicate significant differences. p < 0.05.
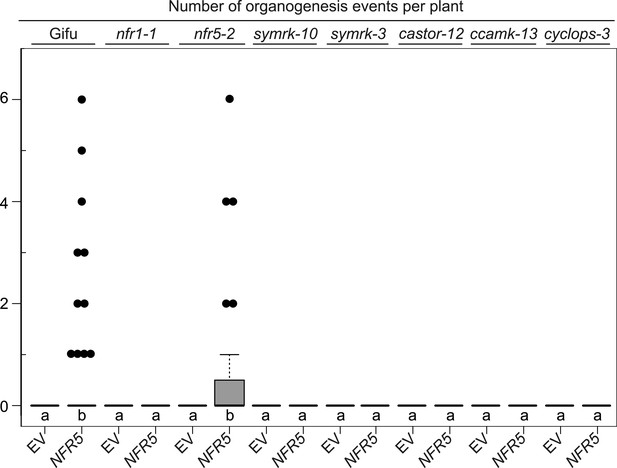
NFR5-mediated spontaneous organogenesis events in Gifu wild-type, nfr1-1, nfr5-2, and common symbiosis mutants.
Hairy roots of L. japonicus Gifu wild-type and different symbiosis defective mutants transformed with the empty vector (EV) or pUB:NFR5-mOrange (NFR5) were generated. Plot represents the number of organogenesis events (nodules and nodule primordia) per plant formed in the absence of rhizobia at 60 dpt. Black dots, data points outside 1.5 IQR of the upper quartile; bold black line, median; box, IQR; whiskers, lowest/highest data point within 1.5 IQR of the lower/upper quartile. A Kruskal–Wallis test followed by false discovery rate correction was performed. Different letters indicate significant differences. p < 0.05.
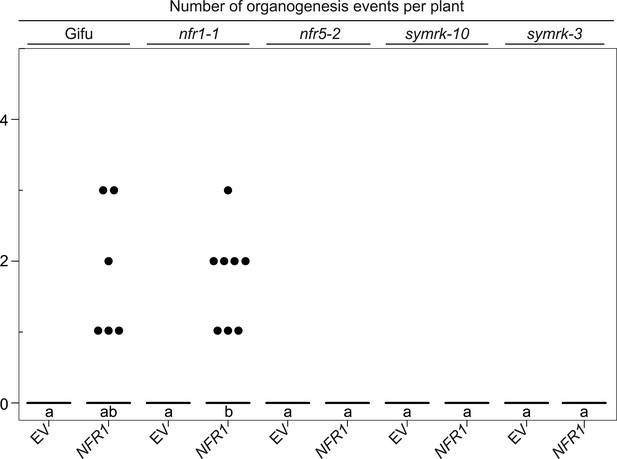
NFR1-mediated spontaneous organogenesis events in Gifu wild-type, nfr1-1, nfr5-2, symrk-10, and symrk-3.
Hairy roots of L. japonicus Gifu wild-type and different symbiosis defective mutants transformed with the empty vector (EV) or pUB:NFR1-mOrange (NFR1) were generated. Plot represents the number of organogenesis events (nodules and nodule primordia) per plant formed in the absence of rhizobia at 60 dpt. Black dots, data points outside 1.5 IQR of the upper quartile; bold black line, median. A Kruskal–Wallis test followed by false discovery rate correction was performed. Different letters indicate significant differences. p < 0.05.
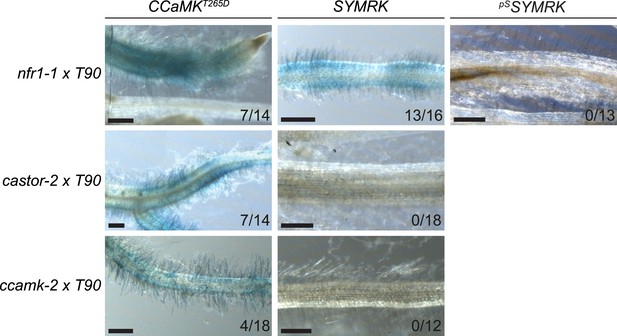
SYMRK-mediated activation of the symbiosis-specific T90 reporter in symbiosis-defective mutants.
Hairy roots of three stable transgenic L. japonicus Gifu reporter lines homozygous for the T90 reporter fusion and the indicated mutant alleles transformed with pUB:CCaMK T265D (CCaMK T265D , a deregulated version of CCaMK), pUB:SYMRK-mOrange (SYMRK), or pSYMRK:SYMRK-RFP ( pS SYMRK) were generated and kept on agar plates for a total of 38 dpt (see ‘Materials and methods’). The vast majority of transgenic root systems did not develop spontaneous nodules at this time point under these growth conditions. GUS activity was analysed by histochemical staining with X-Gluc at 38 dpt. Representative root sections are shown. Number of plants with detectable GUS activity per total plants is indicated. Bars, 500 μm.
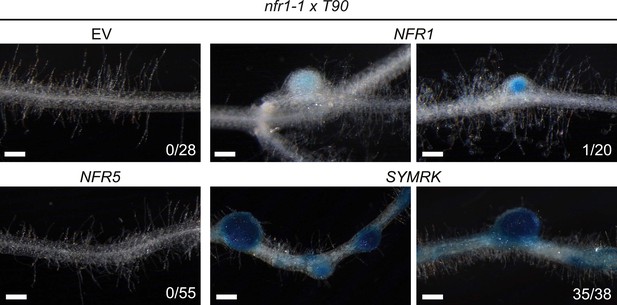
NFR-mediated activation of the symbiosis-specific T90 reporter in the nfr1-1 mutant background.
Hairy roots a stable transgenic L. japonicus Gifu reporter line homozygous for the T90 reporter fusion and the nfr1-1 mutant allele transformed with the empty vector (EV), pUB:NFR1-mOrange (NFR1), pUB:NFR5-mOrange (NFR5), or pUB:SYMRK-mOrange (SYMRK) were generated. GUS activity was analysed by histochemical staining with X-Gluc at 60 dpt. Representative root sections are shown. Number of plants with detectable GUS activity per total number of plants is indicated. Bars, 500 μm.
Additional files
-
Supplementary file 1
(A) Constructs. (B) Oligonucleotides.
- https://doi.org/10.7554/eLife.03891.018





