The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA
Figures
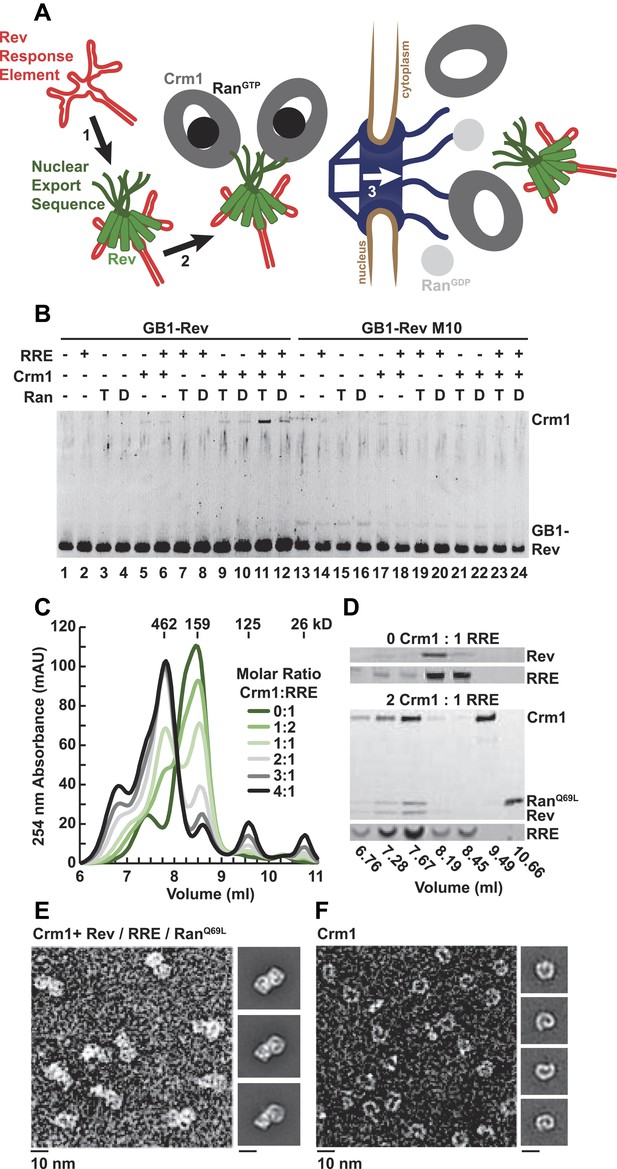
A dimer of Crm1-RanGTP binds the Rev-RRE complex.
(A) Rev assembles on the Rev Response Element (RRE) (step 1) and presents nuclear export sequences (NESs) for Crm1 to recognize in conjunction with RanGTP (step 2). The assembled particle passes through the nuclear pore complex (step 3) to the cytoplasm where Ran GTP hydrolysis stimulates the dissociation of Rev-RRE from Crm1. (B) The gel shows the protein composition from affinity purifications of GB1-Rev incubated in the presence (+) or absence (−) of the RRE, Crm1, and Ran. T or D denotes Ran exchanged with GTPγS or GDP, respectively. RevM10 is a mutation within the NES of Leu78Asp and Glu79Leu (Malim et al., 1989). The gel is representative of three independent experiments. (C–D) Reconstituted Rev-RRE particles preferentially bind two copies of Crm1 and RanGTP. (C) Size-exclusion chromatograms of Rev-RRE with increasing molar ratios of Crm1 and RanQ69L show saturation at a molar ratio of two Crm1 per RRE. Excess Crm1 eluted as free monomers (9.49 ml fraction) or in a small population of larger complexes (6.76 and 7.02 ml fractions). Molecular masses were determined using multi-angle laser light scattering after size-exclusion chromatography and have a technical error of three percent. (D) The composition of peak fractions was analyzed by SDS-PAGE for proteins and by Urea-PAGE for RNA. These results are representative of two independent experiments. (E–F) Negative-stain electron micrographs show two copies of Crm1 in complexes reconstituted with Rev-RRE and RanGTP. Representative fields and class averages of 7196 nuclear export particles (E) and 6445 Crm1 molecules alone (F) are shown.
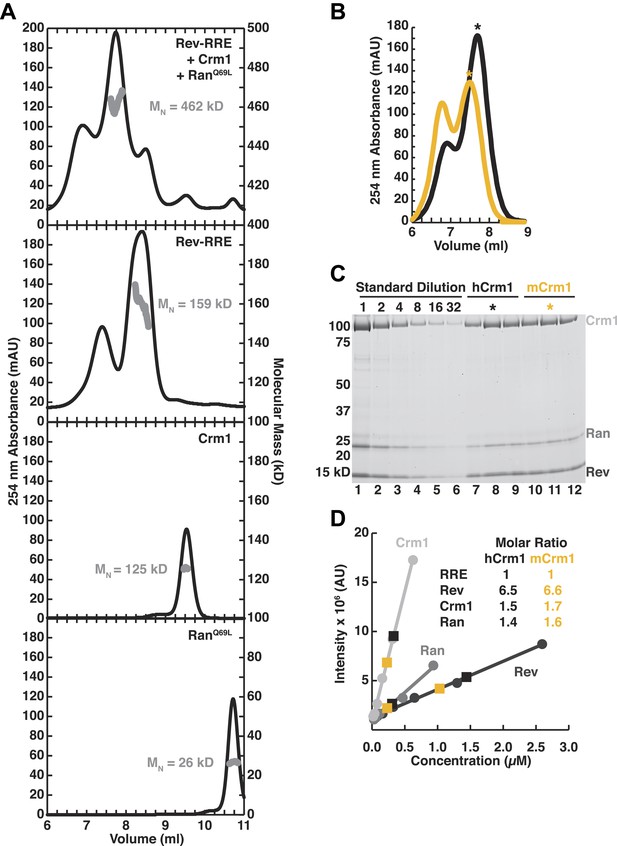
Assessment of the stoichiometry of Crm1, RanGTP, Rev, and the RRE.
(A) Size-exclusion chromatograms of the reconstituted complex (top), Rev-RRE (second from top), Crm1 (third from top), or RanGTP (bottom) with indicated molecular masses (MW) determined by multi-angle laser light scattering after size exclusion chromatography. The RRE has a mass of 80.172 kD and each Rev subunit has a mass of 13.179 kD, so the total 159 kD mass of the complex corresponds to six Rev subunits per RRE. (B–C) Determination of stoichiometry based on protein standards. (B) Size-exclusion chromatograms from Figure 2E of human or murine Crm1 assembled with Rev-RRE and RanGTP with fractions used for determining stoichiometry marked with an asterisk. (C) An SDS-PAGE gel stained with Sypro-Ruby showing a dilution series of protein standards, which were quantified by amino acid analysis, and indicated fractions from the chromatograms in panel B. (D) Quantification of (C) and the calculated stoichiometry of the complex. The RRE concentration was determined by UV spectrometry and used to determine the molar ratios for proteins. These data indicate that this reconstitution had slightly sub-stoichiometric amounts of Crm1 and RanGTPand six subunits of Rev bound to the RRE, with a small population having slightly more Rev bound.
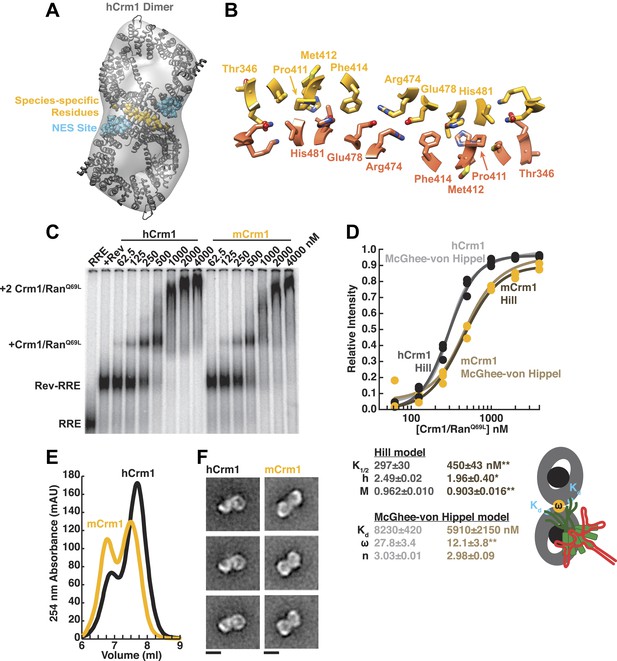
Species-specific residues define a Crm1 dimerization surface that enhances Rev-RRE binding.
(A) A dimer of human Crm1 extracted from a unit cell of a Crm1-Snurportin1 complex that lacks RanGTP (PDB 3GB8) was fit into the EM reconstruction (grey envelope) of Crm1 and RanGTP bound to Rev-RRE (Figure 2—figure supplement 2). Residues that differ between murine and human Crm1 that enhance Rev-RRE activity (Sherer et al., 2011; Elinav et al., 2012) are shown in gold and form an interface between two Crm1 monomers (detailed in panel B and Figure 2—figure supplement 3). Two NES binding sites poised to engage two Rev NES peptides are shown in cyan. (C–D) Human Crm1 recognizes Rev-RRE with higher affinity and increased cooperativity compared to its murine ortholog, shown by gel mobility shift assays with radiolabeled RRE, a molar excess of Rev, and increasing concentrations of Crm1-RanGTP. The Hill equation or a version of the McGhee-von Hippel model (Epstein, 1978) was fit to the quantified data from three independent experiments (R2 > 0.99 for all models compared to average values) as shown in (D) and described in ‘Materials and methods’. Asterisks denote *p < 0.05 and **p < 0.01 from one-tailed t-tests. A schematic model of the complex is shown to highlight the physical meaning of the McGhee-von Hippel parameters. (E) Size-exclusion chromatograms of Rev-RRE complexes assembled with murine Crm1 or human Crm1 show that murine Crm1 elutes with a larger apparent mass, presumably due to a larger hydrodynamic radius in the absence of Crm1-Crm1 interactions. (F) Class averages of Rev-RRE complexes bound to a dimer of human (426 particles) or murine (617 particles) Crm1 from negatively-stained micrographs of particles from (E). Murine Crm1 particles showed a wider range of sizes than the human particles, with most being larger, and the murine particles showed different features, such as an hourglass shape with fewer details in the center of the particle than human Crm1 particles. Scale bars show 10 nm.
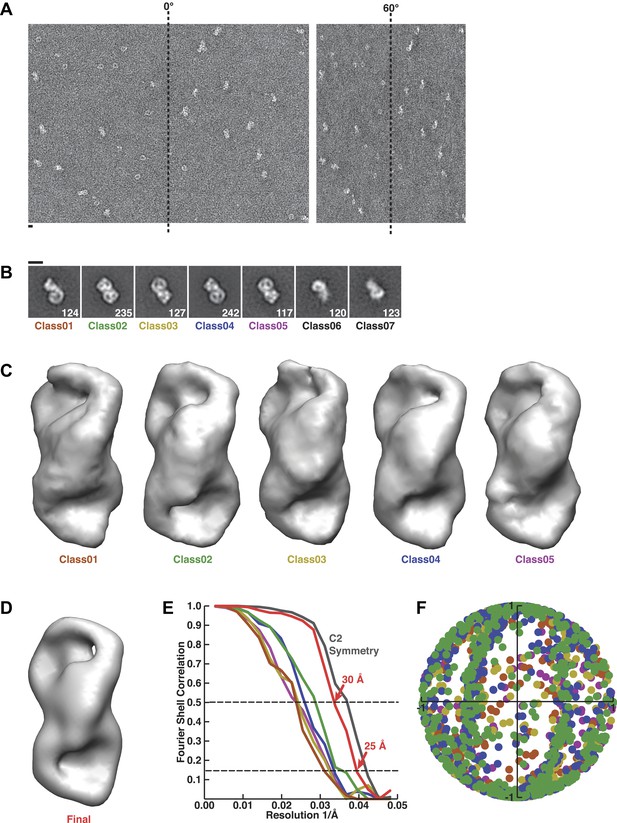
Random conical tilt reconstruction and refinement.
(A) Representative fields from a random conical tilt pair of particles assembled with Rev-RRE, Crm1, and RanGTP. (B) Class averages from untilted particles used for random conical tilt. Particle numbers are inset in each image. Classes six and seven were observed with lower concentrations of sample and resemble partially disassembled complexes. Scale bars show 10 nm. (C) Volumes from each class after random conical tilt refinement show similar features among all classes suggesting the classes are different views of the same particle. The volume from the final reconstruction is shown in (D). (E) Fourier shell correlation curves for each random conical tilt class and the final reconstruction. The boost in Fourier shell correlation when C2 symmetry is applied during refinement confirms the symmetric arrangement of bowl-like densities. (F) Angular distribution from projection matching shows no major gap in Euler space.
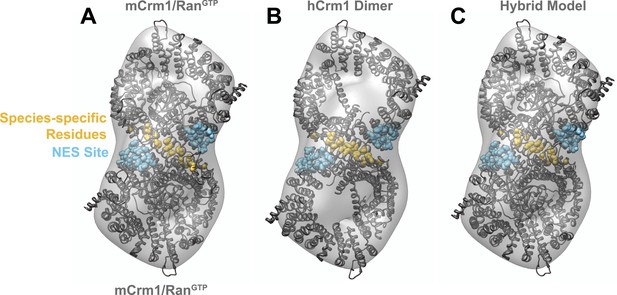
Comparison of fitting murine Crm1 or human Crm1 crystal structures in the EM reconstruction.
(A) Two murine Crm1-RanGTP (PDB 3NC0) monomers individually fit well (Correlation (R) = 0.7526 or 0.7393) into two bowl-like densities from the reconstruction of Crm1-RanGTP bound to Rev-RRE. The final fit places residues different between murine and human Crm1 (gold) to potentially form an interface between two Crm1 monomers. (B) From the fit of murine Crm1, we inferred that the same residues might form a crystal contact between Crm1 monomers in the crystal structure of human Crm1 bound to Snurportin-1 (PDB 3GB8) and this, indeed, was observed. The fit of the human Crm1 dimer structure is shown (R = 0.8227). Although the crystal structures of murine Crm1 either have two Crm1 molecules in the asymmetric unit or other dimers in the unit cell, these alternate dimers use different crystal contacts to build the unit cell and do not have the same conformation as the human Crm1 dimer (data not shown). (C) A hybrid model of the Crm1 dimer bound to RanGTP. The human Crm1 interface was used to orient two copies of murine Crm1 bound to RanGTP by aligning murine Crm1 with the human Crm1 interface, which shows little deviation between Cα atoms. The final model fits well (R = 0.9047) into the EM map and was used to search for Rev-RRE density shown in Figure 3.
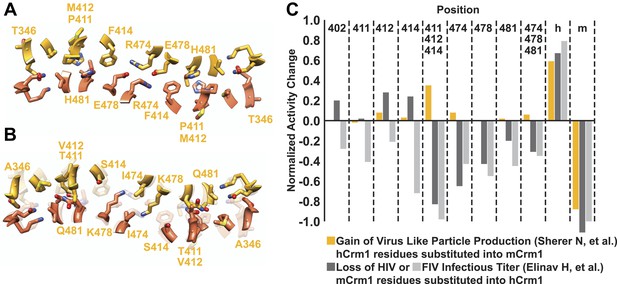
A comparison of species-specific residues at the interface of the Crm1 dimer.
Species-specific residues form many interactions in the human Crm1 dimer interface (A) but few at the analogous murine positions (B). Aligning the Cα atoms from murine and human Crm1 at the dimer interface (B) shows steric clashes between side chains near Thr411 in murine Crm1 interface and between the side chains of His352 and Thr487 that would prevent murine Crm1 dimerization. (C) Swapping residues between human and murine Crm1 in the dimer interface alters virus production. A summary of data from references (Sherer et al., 2011) and (Elinav et al., 2012) testing the effect of murine or human residues on virus-like particle production or infectivity of HIV or FIV vectors shows the biological relevance of the Crm1 interface. Sherer et al. substituted human residues into murine Crm1 to determine which residues enhanced virus-like particle production (Sherer et al., 2011). Elinav et al. substituted mouse residues into human Crm1 and scored for loss of function (Elinav et al., 2012). In both studies, the gain in activity denoted by ‘h’ is a substitution of all human interface residues into murine Crm1 and vice versa for the loss of activity denoted by ‘m’. Data were normalized according to the following equations: Gain of Function = (mutant activity−murine Crm1 activity)/(human Crm1 activity−murine Crm1 activity) and Loss of Function = Gain of function−1.
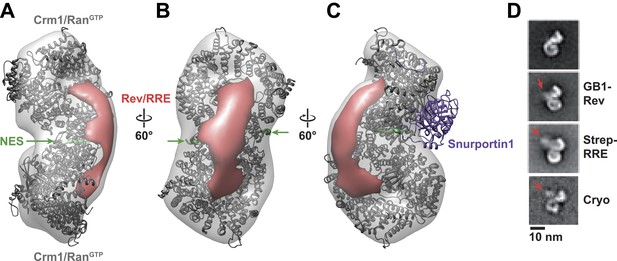
Rev-RRE binds between NES binding sites on the Crm1 dimer.
(A–B) A model of the Crm1-RanGTP dimer displays extra density (red) on one side of the structure that locates the Rev-RRE complex between the Rev NESs bound to each Crm1 (green). The Crm1-RanGTP dimer model was built by aligning two murine Crm1-RanGTP complexes onto the human Crm1 dimer. (C) A rotated view of the structure showing the location of Snurportin1 bound to a Crm1 monomer on the opposite side of the protein. (D) Class averages of particles containing Rev tagged with GB1 or RRE tagged with streptavidin, and cryo-EM of the complex without any tags, confirm the location of Rev-RRE. Raw micrographs, class averages, and particle numbers are shown in Figure 3—figure supplement 2.
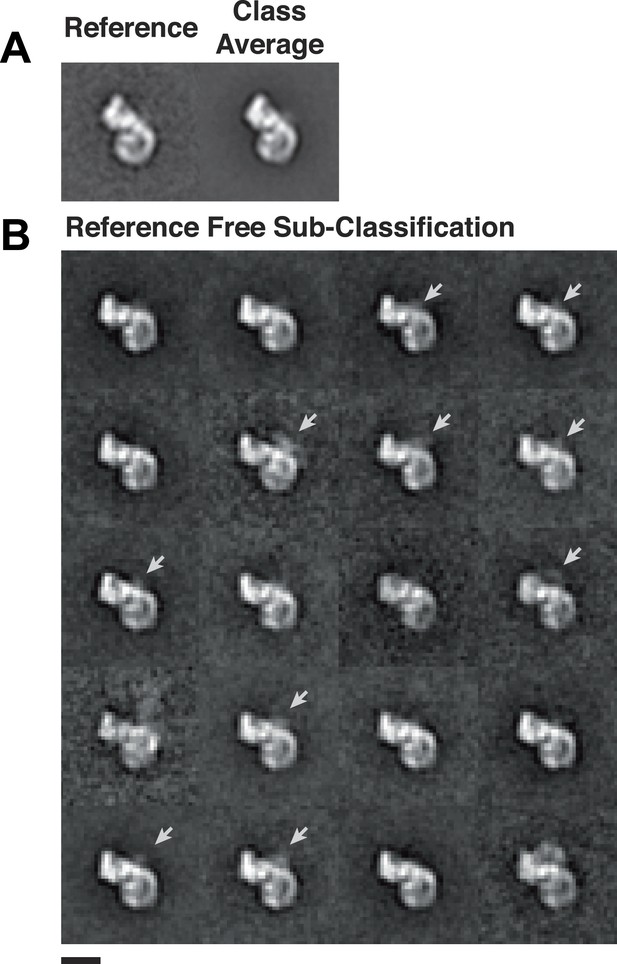
Heterogeneity within class averages reveals Rev-RRE density.
Particles collected from an independent data set were aligned to references from random conical tilt class averages (A), and 1150 particles from one class were sub-classified using a reference-free alignment (B). Some of the sub-classes further reveal extra densities (arrows) at the Rev-RRE location. Scale bars show 10 nm.
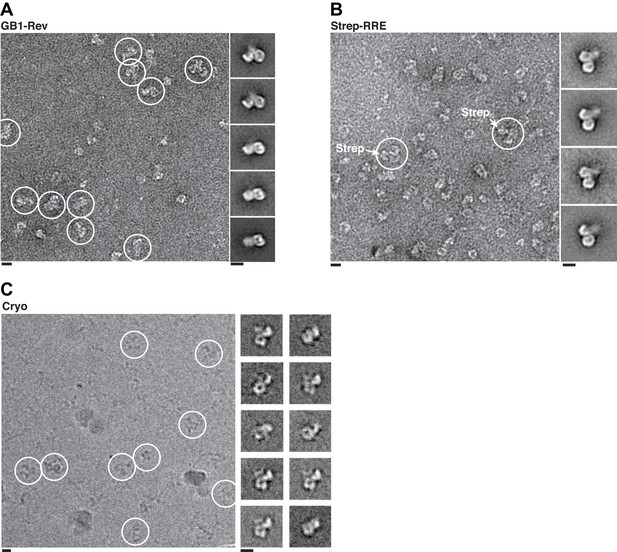
Localization of Rev-RRE density.
Representative fields and class averages from negative stain (A–B) or cryo-electron micrographs (C) show extra density from Rev-RRE when Rev is tagged with GB1 (A, 794 particles) or the RRE tagged with Streptavidin (B, 383 particles). Extra density from Rev-RRE also is apparent in cryo-EM images of complexes that do not have any additional tag (C, 3262 particles), where the contrast is generated from the particles themselves rather than from uranyl ions that typically obscure nucleic acids. Scale bars show 10 nm.
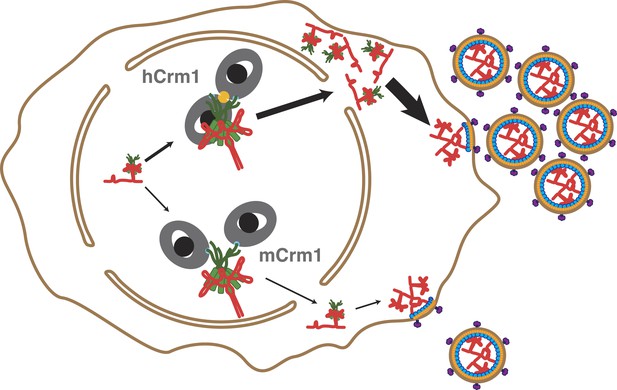
Cooperative assembly of a nuclear export complex with human Crm1 enhances nuclear export and virion production.
Human Crm1 (top) associates with Rev-RRE with higher affinity than murine Crm1 (bottom) due to species-specific residues that form the dimer interface (gold). This increased association amplifies into even larger differences in nuclear export activity and virion assembly between the two Crm1s.
Additional files
-
Supplementary file 1
Plasmids for protein and RNA expression.
- https://doi.org/10.7554/eLife.04121.013
-
Supplementary file 2
Primers.
- https://doi.org/10.7554/eLife.04121.014





