Endocrine remodelling of the adult intestine sustains reproduction in Drosophila
Figures
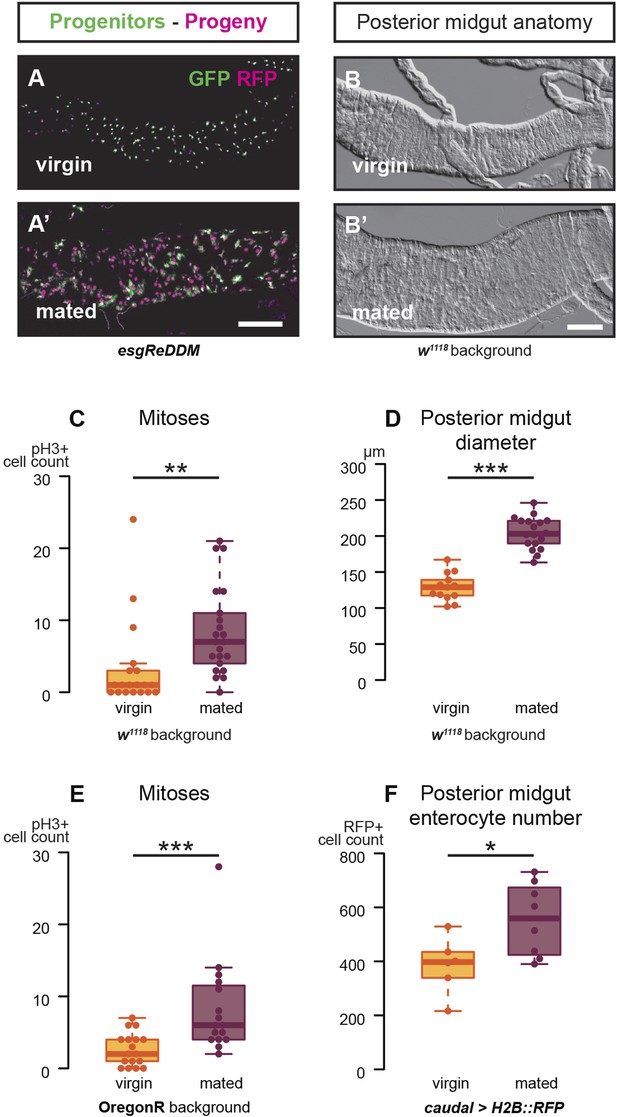
Mating increases ISC proliferation and gut size.
(A, A′) Using the esgReDDM tracer (Antonello et al., 2015), intestinal progenitors (intestinal stem cells (ISCs) and enteroblasts) are labelled with GFP and RFP, whereas the postmitotic progeny (enterocytes (ECs) and enteroendocrine cells) that these progenitors give rise to in a defined time window is labelled with RFP only (see Supplemental Information for additional details). At 3 days after mating, the posterior midgut of mated flies contains more newly generated postmitotic progeny (A) compared to age-matched virgins (A′). It has also become visibly larger (B, B′). At this time point, these guts also have a higher number of nuclei marked by the mitotic marker pH3 in both w1118 and OregonR backgrounds (C, p = 0.008, and E, p < 0.001, negative binomial GLM), although the proliferation increase is transient (data not shown). The size increase is quantified in the posterior midgut by measuring midgut diameter (D, p < 0.001, t-test) and counting the number of cells labelled by the EC marker caudal-Gal4 (F, p = 0.02, t-test). See Table 1 for full genotypes.
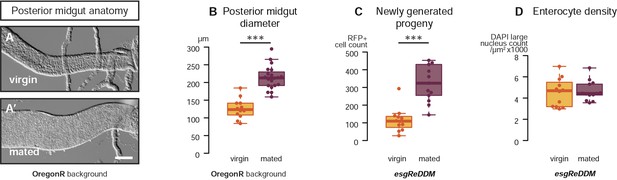
Mating re-sizes the Drosophila gut.
The increase in gut size at 3 days after mating is also measurable (A, A′) and significant (B, p < 0.001, t-test) in the OregonR background. The esgReDDM tracing system reveals that mated guts contain more cells generated in the last 7 days if the fly had been mated in that time (C, p < 0.001, t-test) than if it had not. The size increase is not due to stretching of the tissue, as the density of nuclei in the posterior midgut remains the same (D, p = 0.77, t-test). See Table 1 for full genotypes.
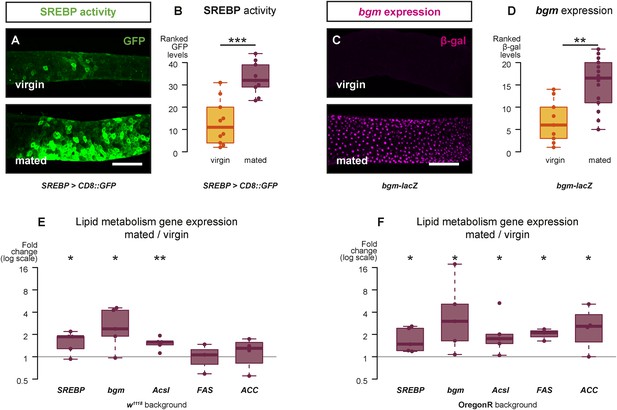
Mating changes the activity and/or expression of lipid metabolism genes in the intestine.
At 3 days after mating, increased expression of a reporter that replicates the transcriptional regulation and post-translational modification of sterol regulatory element-binding protein (SREBP) is apparent in the posterior midgut (A, A′, quantified in B, p < 0.001, Mann–Whitney test). A bgm transcriptional reporter is also increased specifically in the ECs of the posterior midgut following mating (C, C′, quantified in D, p = 0.002, Mann–Whitney test). Transcript abundance of SREBP, bgm, and the SREBP targets Acyl-CoA synthetase long-chain (Acsl), Fatty acid synthase (FAS), and Acetyl-CoA carboxylase (ACC) is increased by mating in either one or both of the w1118 and OregonR backgrounds (E w1118: p = 0.02 SREBP, p = 0.02 bgm, p = 0.005 Acsl, p = 0.5 FAS, p = 0.3 ACC; F OregonR: p = 0.02 SREBP, p = 0.03 bgm, p = 0.03 Acsl, p = 0.01 FAS, p = 0.04 ACC, paired one-tailed t-test). See Table 1 for full genotypes.

Systemic JH secreted after mating acts directly in the intestinal epithelium to drive reproductive remodelling.
Circulating juvenile hormone (JH) is elevated after mating in the haemolymph of female flies (A, p = 0.02 at 24 hr, p = 0.002 at 48 hr, t-test with Holm's correction). Increased tissue renewal (B, B′) and SREBP activation (C, C′, quantified in D, p < 0.001, Mann–Whitney test) are apparent following a 3-day dietary supplementation with JH analogue (JHa). JHa treatment is sufficient to increase mitoses (E, p < 0.001, negative binomial GLM) and size (H, p < 0.001, t-test) of the posterior midgut. Conversely, when the endogenous JH source is genetically ablated by means of Aug21 > NiPp1 (Yamamoto et al., 2013), the proliferation and size increase that follow mating are abolished, although they can be reinstated by feeding JHa (proliferation F, p < 0.001 between Aug21/+ and Aug21 > NiPp1 mated, p < 0.001 between Aug21 > NiPp1 and NiPp1/+ mated, p < 0.001 between Aug21 > NiPp1 and Aug21 > NiPp1 + JHa mated; all relevant comparisons between virgins are not significant, negative binomial GLM with Holm's correction; gut diameter I, p = 0.002 between Aug21/+ and Aug21 > NiPp1 mated, p < 0.001 between Aug21 > NiPp1 and NiPp1/+ mated, p < 0.001 between Aug21 > NiPp1 and Aug21 > NiPp1 + JHa mated; all relevant comparisons between virgins are not significant, t-test with Holm's correction). Downregulation of either gce or Met in adult progenitors abrogates post-mating proliferation (G, p < 0.001 between esgReDDM/+ and esgReDDM > gce RNAi mated, p < 0.001 between esgReDDM/+ and esgReDDM > Met RNAi mated, negative binomial GLM with Holm's correction) and gut size increase (J, p < 0.001 between esgReDDM/+ and esgReDDM > gce RNAi mated, p < 0.001 between esgReDDM/+ and esgReDDM > Met RNAi mated, t-test with Holm's correction). The upregulation of bgm reporter upon mating is abolished by the downregulation of gce, but not Met, in ECs using the EC-specific driver Mex-Gal4 (K–K′′′′). See Table 1 for full genotypes.
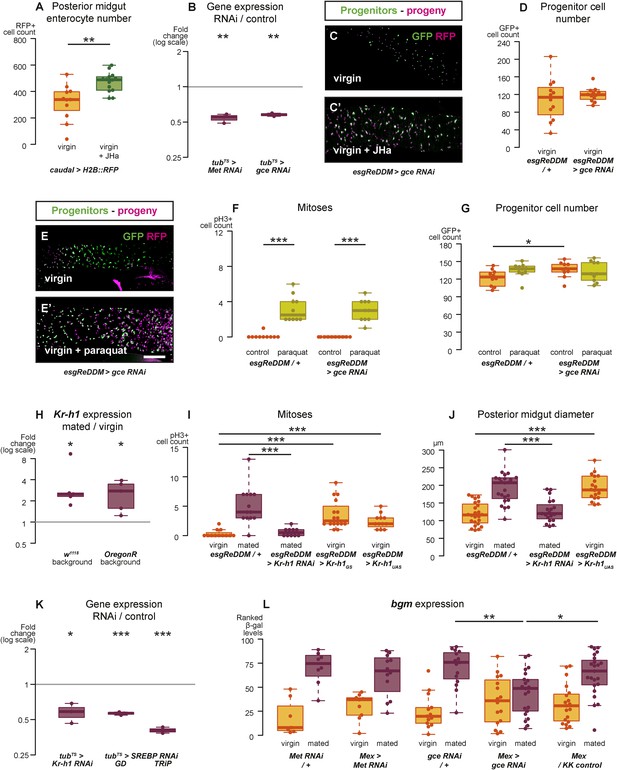
Intestinal JH signalling is relayed through Kr-h1 and underlies mating-dependent intestinal growth and gene expression phenotypes.
JHa application in virgin females results in a net growth of the gut, as shown by the increase in caudal-marked cells (A, p = 0.004, t-test). The JH signalling pathway can be targeted using RNAi constructs against the receptors Met and gce, which decrease transcript abundance compared to a tubts control when expressed globally in larvae for 3 hr at 29°C (B, p = 0.002 for Met, p = 0.005 for gce, paired one-tailed t-test). Consequently, using esgReDDM to specifically knockdown gce in adult intestinal progenitor cells abolishes the proliferative effect of JHa application (C, C′), but does not reduce the number of progenitors after 7 days of downregulation (D, p > 0.05, t test). Progenitors in which gce is downregulated can still proliferate normally to replenish a gut damaged by a 24 hr application of the toxin paraquat (E, E′ with quantification of mitoses in F, p < 0.001 for both esgReDDM/+ and esgReDDM > gce RNAi, p > 0.05 for all other relevant comparisons, t test with Holm's correction) and the number of progenitors is not reduced by this treatment (G, p = 0.04 between esgReDDM/+ untreated control and esgReDDM > gce RNAi untreated control, p > 0.05 for all other relevant comparisons, t test with Holm's correction). The transcription factor Kruppel homolog 1 (Kr-h1), a well-established effector of JH responses, is transcriptionally upregulated after 3 days of mating (H, p = 0.02 in w1118, p = 0.02 in OregonR, paired two-tailed t-test). Kr-h1 function is necessary and sufficient for the re-sizing of the gut after mating, as its downregulation in intestinal progenitors through RNA interference using esgReDDM prevents the increase in proliferation (I, p < 0.001 between esgReDDM/+ and esgReDDM > Kr-h1 RNAi mated, negative binomial GLM with Holm's correction) and gut size (J, p < 0.001 between esgReDDM/+ and esgReDDM > Kr-h1 RNAi mated, t-test with Holm's correction) typically observed after 7 days of mating, while overexpression of Kr-h1 constructs from the same cells recapitulates the effect of mating in virgins (proliferation, I, p < 0.001 between esgReDDM/+ and esgReDDM > Kr-h1GS virgin, p < 0.001 between esgReDDM/+ and esgReDDM > Kr-h1UAS virgin, negative binomial GLM with Holm's correction; gut size J, p < 0.001 between esgReDDM/+ and esgReDDM > Kr-h1UAS virgin, t-test with Holm's correction). RNAi constructs against Kr-h1 and SREBP are effective in downregulating these genes; they decrease transcript abundance compared to a tubts control when expressed globally in larvae for 3 hr at 29°C (K, p = 0.02 for Kr-H1, p < 0.001 for both SREBP constructs, paired one-tailed t-test). Downregulating gce constitutively from ECs using Mex-Gal4 significantly suppresses the transcriptional increase of the lipid metabolism gene bgm upon mating, as indicated by the intensity ranking of a gce reporter (L, p = 0.004 between gce RNAi/+ and Mex > gce RNAi, p = 0.02 between Mex > gce RNAi and Mex/KK control, Mann–Whitney test with Holm's correction; relevant comparisons with Met RNAi are not significant). See Table 1 for full genotypes.
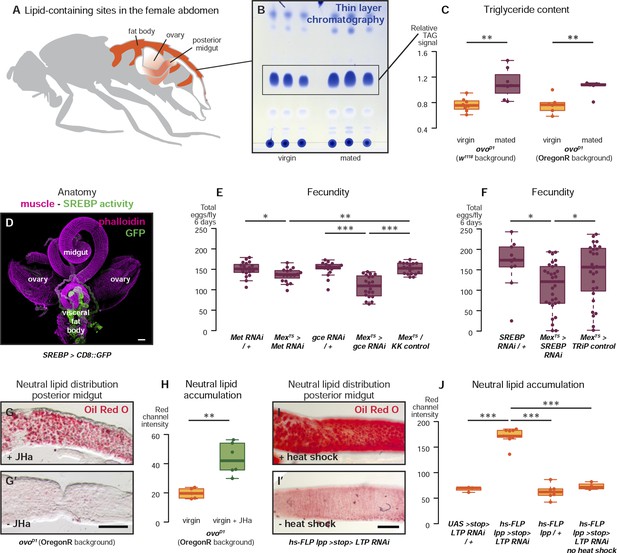
Metabolic remodelling of ECs by JH sustains reproduction.
Lipid-harbouring tissues (fat body, posterior midgut, and ovary) are found in close proximity in the fly's abdomen (represented schematically in A, and in confocal microscopy in D). The amount of stored triglycerides (TAG) in the carcass of 3-day mated sterile female flies is increased compared to virgins (B, p = 0.003 in w1118, p = 0.009 in OregonR, t-test), as quantified by thin-layer chromatography (C). Adult-specific downregulation of JH receptors gce and Met or SREBP in ECs reduces the total progeny produced by females in the 6 days following their first mating (E, p = 0.01 between Met RNAi/+ and Mexts > Met RNAi, p = 0.007 between Mexts > Met RNAi and Mexts/KK control, p < 0.001 between gce RNAi/+ and Mexts > gce RNAi, p = 0.007 between Mexts> gce RNAi and Mexts/KK control; F p = 0.04 between SREBP RNAi/+ and Mexts > SREBP RNAi, p = 0.04 between Mexts > SREBP RNAi and Mexts/TRiP control, t-test with Holm's correction). In the absence of the ovarian lipid sink in sterile ovoD1 virgin flies, treatment with JHa increases neutral lipid content, as revealed by Oil Red O staining, in the posterior midgut (G, G′, quantified in H: p = 0.002, t-test). Acute block of lipid export by heat-shock activation of lpp > stop > LTP RNAi (Palm et al., 2012) in virgin females results in heavy accumulation of neutral lipid in this gut region, further indicating that this midgut region provides a net source of lipid in adult flies (I, quantified in J: p < 0.001 between LTP RNAi/+ and lpp > stop > LTP RNAi, p < 0.001 between lpp > stop > LTP RNAi and lpp > stop>/+, p < 0.001 between lpp > stop > LTP RNAi and lpp > stop > LTP RNAi heat shock control, t-test with Holm's correction). See Table 1 for full genotypes.
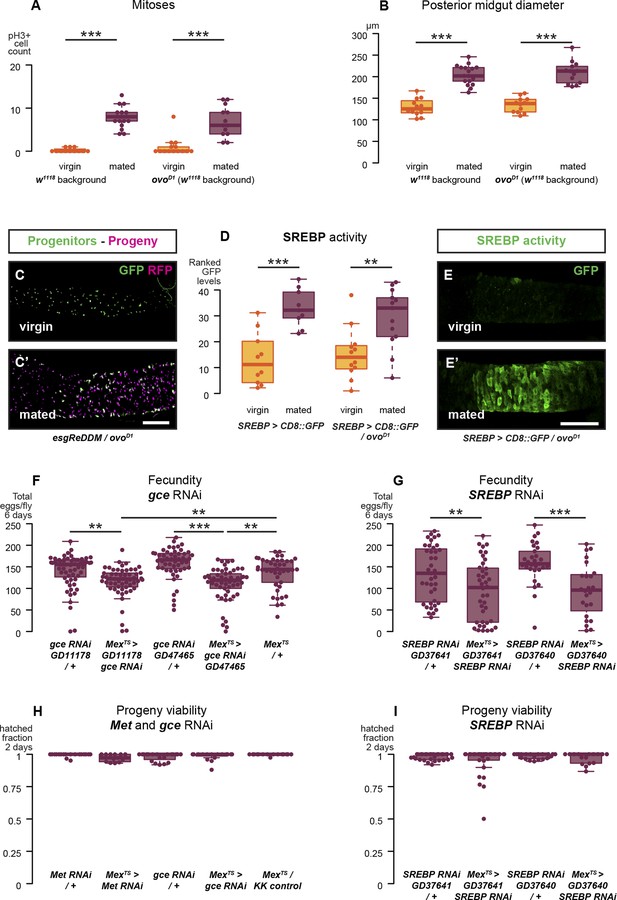
Reproductive intestinal remodelling is uncoupled from germline demands and is needed to sustain reproduction.
Sterile females carrying the ovoD1 mutation experience the post-mating increase in progenitor proliferation (A, p < 0.001 for w1118 and ovoD1, negative binomial GLM, visualised in C and C′ using the esgReDDM tracing system), gut size increase (B, p < 0.001 for w1118 and ovoD1, t-test), and SREBP reporter activation (D, p < 0.001 for w1118 and p = 0.005 for ovoD1, Mann–Whitney test, visualised in E and E′). The role of intestinal remodelling in enhancing reproductive capacity is confirmed with additional RNA interference lines against the JH receptor gce (chosen because of its larger effect in Figure 4E; F, p = 0.002 between GD11178/+ and Mexts> GD11178, p = 0.008 between Mexts > GD11178 and Mexts/+, p < 0.001 between GD47465/+ and Mexts> GD47465, p = 0.003 between Mexts > GD11178 and Mexts/+, t-test with Holm's correction) and SREBP (G, p = 0.008 between GD37641/+ and Mexts > GD37641, p < 0.001 between GD37640/+ and Mexts > GD37640, t-test). Despite these effect on fecundity, eggs laid by gce, Met, or SREBP RNAi mothers are viable (H and I, mean hatched fraction >0.9 for all groups, p > 0.05 for all relevant comparisons, t-test). See Table 1 for full genotypes.
Tables
Full genotypes
| Genotype in text/figure | Full genotype | Figure panel(s) |
|---|---|---|
| esgReDDM | w; esg-Gal4, UAS-mCD8::GFP/+; tub-Gal80ts, UAS-H2B::RFP/+; + | Figure 1A, Figure 1—figure supplement 1C,D, Figure 3B |
| w1118 background | w1118; +; +; + | Figure 1B–D, Figure 2E, Figure 3E,H, Figure 3—figure supplement 1H, Figure 4—figure supplement 1A,B |
| OregonR background | +; +; +; + | Figure 1E, Figure 1—figure supplement 1A,B, Figure 2F, Figure 3—figure supplement 1H |
| caudal > H2B::RFP | w; caudal-Gal4/+; UAS-H2B::RFP/+; + | Figure 1F, Figure 3—figure supplement 1A |
| SREBP > CD8::GFP | w/+; SREBP-Gal4/+; UAS-CD8::GFP/+; + | Figure 2A,B, Figure 3C,D, Figure 4D, Figure 4—figure supplement 1D |
| bgm-lacZ | w/+; bgm-lacZ/+; +; + | Figure 2C,D |
| CantonS background | +; +; +; + | Figure 3A |
| Aug21/+ | w; Aug21-Gal4/+; +; + | Figure 3F,I |
| Aug21 > NiPp1 | w; Aug21-Gal4/+; UAS-NiPp1/+; + | Figure 3F,I |
| NiPp1/+ | w; +; UAS-NiPp1/+; + | Figure 3F,I |
| esgReDDM/+ | w; esg-Gal4, UAS-mCD8::GFP/+; tub-Gal80ts, UAS-H2B::RFP/+; + | Figure 3G,J, Figure 3—figure supplement 1D,F,G,I,J |
| esgReDDM > gce RNAi | w; esg-Gal4, UAS-mCD8::GFP/UAS-gce RNAi KK101814; tub-Gal80ts, UAS-H2B::RFP/+; + | Figure 3G,J, Figure 3—figure supplement 1C–G |
| esgReDDM > Met RNAi | w; esg-Gal4, UAS-mCD8::GFP/UAS-Met RNAi KK100638; tub-Gal80ts, UAS-H2B::RFP/+; + | Figure 3G,J |
| esgReDDM > Kr-h1 RNAi | w; esg-Gal4, UAS-mCD8::GFP/UAS-Kr-h1 RNAi KK107935; tub-Gal80ts, UAS-H2B::RFP/+; + | Figure 3—figure supplement 1I,J |
| esgReDDM > Kr-h1GS | w; esg-Gal4, UAS-mCD8::GFP/UAS-Kr-h1GS;tub-Gal80ts, UAS-H2B::RFP/+; + | Figure 3—figure supplement 1I |
| esgReDDM > Kr-h1UAS | w; esg-Gal4, UAS-mCD8::GFP/UAS-Kr-h1UAS; tub-Gal80ts, UAS-H2B::RFP/+; + | Figure 3—figure supplement 1I,J |
| tubts> Met RNAi | w; tub-Gal80ts/UAS-Met RNAi KK100638; tub-Gal4, UAS-mCD8::GFP/+; + | Figure 3—figure supplement 1B |
| tubts > gce RNAi | w; tub-Gal80ts/UAS-gce RNAi KK101814; tub-Gal4, UAS-mCD8::GFP/+; + | Figure 3—figure supplement 1B |
| tubts > Kr-h1 RNAi | w; tub-Gal80ts/UAS-Kr-h1 RNAi KK107935; tub-Gal4, UAS-mCD8::GFP/+; + | Figure 3—figure supplement 1K |
| tubts > SREBP RNAi GD | w; tub-Gal80ts/UAS-SREBP RNAi GD37640; tub-Gal4, UAS-mCD8::GFP/+; + | Figure 3—figure supplement 1K |
| tubts > SREBP RNAi TRiP | w; tub-Gal80ts +; tub-Gal4, UAS-mCD8::GFP/UAS-SREBP RNAi TRiP34073; + | Figure 3—figure supplement 1K |
| tubts/+ | w; tub-Gal80ts/+; tub-Gal4, UAS-mCD8::GFP/+; + | Figure 3—figure supplement 1B,K (control) |
| Met RNAi/+ | w; bgm-lacZ/UAS-Met RNAi KK100638; +; + | Figure 3K, Figure 3—figure supplement 1L |
| Mex > Met RNAi | w; Mex-Gal4, bgm-lacZ/UAS-Met RNAi KK100638; +; + | Figure 3K, Figure 3—figure supplement 1L |
| gce RNAi/+ | w; bgm-lacZ/UAS-gce RNAi KK101814; +; + | Figure 3K, Figure 3—figure supplement 1L |
| Mex > gce RNAi | w; Mex-Gal4, bgm-lacZ/UAS-gce RNAi KK101814; +; + | Figure 3K, Figure 3—figure supplement 1L |
| Mex/KK control | w; Mex-Gal4, bgm-lacZ/attp40; +; + | Figure 3K, Figure 3—figure supplement 1L |
| ovoD1 (w1118 background) | w1118; +; ovoD1/+; + | Figure 4C, Figure 4—figure supplement 1A,B |
| ovoD1 (OregonR background) | +/w1118; +; ovoD1/+; + | Figure 4C,G,H |
| Met RNAi/+ | w; UAS-Met RNAi KK100638/+; +; + | Figure 4E, Figure 4—figure supplement 1H |
| Mexts> Met RNAi | w; Mex-Gal4/UAS-Met RNAi KK100638; tub-Gal80ts/+; + | Figure 4E, Figure 4—figure supplement 1H |
| gce RNAi/+ | w; UAS-gce RNAi KK101814/+; +; + | Figure 4E, Figure 4—figure supplement 1H |
| Mexts > gce RNAi | w; Mex-Gal4/UAS-gce RNAi KK101814; tub-Gal80ts/+; + | Figure 4E, Figure 4—figure supplement 1H |
| Mexts/KK control | w; Mex-Gal4/attp40; tub-Gal80ts/+; + | Figure 4E, Figure 4—figure supplement 1H |
| SREBP RNAi/+ | w/y, v; +; UAS-SREBP RNAi 34073; + | Figure 4F |
| Mexts> SREBP RNAi | w/y, v; Mex-Gal4/+; tub-Gal80TS/UAS-SREBP RNAi 34073; + | Figure 4F |
| Mexts/TRiP control | w/y, v; Mex-Gal4/+; tub-Gal80ts/UAS-GFP; + | Figure 4F |
| hs-FLP lpp > stop > LTP RNAi | w, hs-FLP/w; lpp-Gal4/+; UAS > stop > LTP RNAi/+; + | Figure 4I,J |
| UAS > stop > LTP RNAi/+ | w; +; UAS > stop > LTP RNAi/+; + | Figure 4J |
| hs-FLP lpp/+ | w, hs-FLP/w; lpp-Gal4/+; +; + | Figure 4J |
| esgReDDM/ovoD1 | ovoD1/w; esg-Gal4, UAS-mCD8::GFP/+; tub-Gal80ts, UAS-H2B::RFP/+; + | Figure 4—figure supplement 1C |
| SREBP > CD8::GFP/ovoD1 | w/+; SREBP-Gal4/+; UAS-CD8::GFP/ovoD1; + | Figure 4—figure supplement 1D,E |
| gce RNAi GD11178/+ | w; UAS-gce RNAi GD11178; +; + | Figure 4—figure supplement 1F |
| Mexts> gce RNAi GD11178 | w; Mex-Gal4/UAS-gce RNAi GD11178; tub-Gal80ts/+; + | Figure 4—figure supplement 1F |
| gce RNAi GD47465/+ | w; +; UAS-gce RNAi GD47465/+; + | Figure 4—figure supplement 1F |
| Mexts> gce RNAi GD47465 | w; Mex-Gal4/+; tub-Gal80ts/UAS-gce RNAi GD47465; + | Figure 4—figure supplement 1F |
| Mexts/+ | w; Mex-Gal4/+; tub-Gal80ts+; + | Figure 4—figure supplement 1F |
| SREBP RNAi GD37641/+ | w; UAS-SREBP RNAi GD37641/+; +; + | Figure 4—figure supplement 1G,I |
| Mexts> GD37641 | w; Mex-Gal4/UAS-SREBP RNAi GD37641; tub-Gal80ts/+; + | Figure 4—figure supplement 1G,I |
| SREBP RNAi GD37640/+ | w; UAS-SREBP RNAi GD37640/+; + | Figure 4—figure supplement 1G,I |
| Mexts> RNAi GD37640 | w; Mex-Gal4/+; tub-Gal80ts/UAS-SREBP RNAi GD37640; + | Figure 4—figure supplement 1G,I |





