Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats
Figures
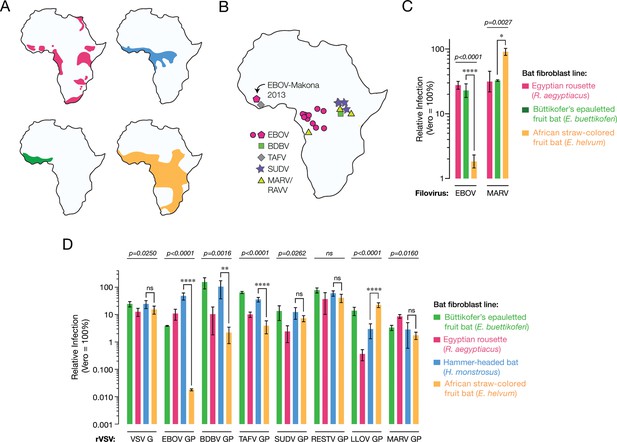
African straw-colored fruit bat cells are refractory to EBOV entry and infection.
(A) Ranges of African pteropodids Rousettus aegyptiacus (pink), Hypsignathus monstrosus (blue), Epomops buettikoferi (green) and Eidolon helvum (yellow) (source: IUCN Redlist). (B) Locations of known filovirus outbreaks. (C) Infection of pteropodid kidney fibroblast cell lines with authentic filoviruses. Means ± standard deviations (n ≥ 3) from two biological replicates are shown. (D) Infections with recombinant vesicular stomatitis viruses (rVSVs) bearing filovirus glycoproteins. BDBV, Bundibugyo virus; TAFV, Taı¨ Forest virus; SUDV, Sudan virus; RESTV, Reston virus; LLOV, Lloviu virus. Means ± SD (n = 3–4) from two biological replicates are shown. In panels C and D, the infectivity of each virus was normalized to that obtained in Vero grivet monkey cells. Means for infection of the different cell lines by each virus were compared by one-way ANOVA (p-value indicated above each group of bars). Tukey’s post hoc test was used to compare infection means on Hypsignathus monstrosus vs Eidolon helvum cells (*p < 0.05; ****p < 0.0001; ns, no statistical significance).
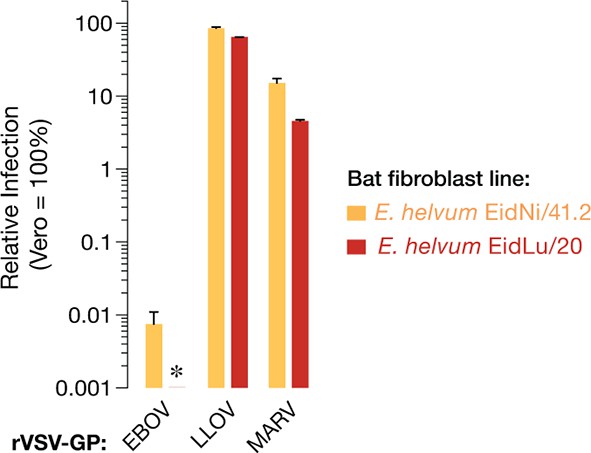
Two additional African straw-colored fruit bat cell lines are selectively refractory to EBOV entry and infection.
Infection of EidNi/41.2 kidney fibroblasts and EidLu/20 lung fibroblasts with recombinant VSV (rVSVs) bearing filovirus glycoproteins. The infectivity of each virus was normalized to that obtained in Vero grivet monkey cells. Asterisk indicates value below the limit of detection of the infection assay. Means ± SD (n = 4) from two biological replicates are shown.
The NPC1-dependent entry and infection block in African straw-colored fruit bat cells is selective for EBOV.
(A) Infection of pteropodid kidney fibroblast cell lines stably expressing human NPC1 (HsNPC1) with authentic filoviruses. (B) Infection of pteropodid kidney fibroblast cell lines with recombinant VSV (rVSVs) bearing filovirus glycoproteins. IU/ml, infectious units per ml. Means ± SD (n = 3) from a representative experiment are shown in each panel. Means for infection of cell lines lacking or ectopically expressing HsNPC1 were compared by unpaired two-tailed Student’s t-test with Welch’s correction (***p< 0.001; ****p< 0.0001; ns, no statistical significance).
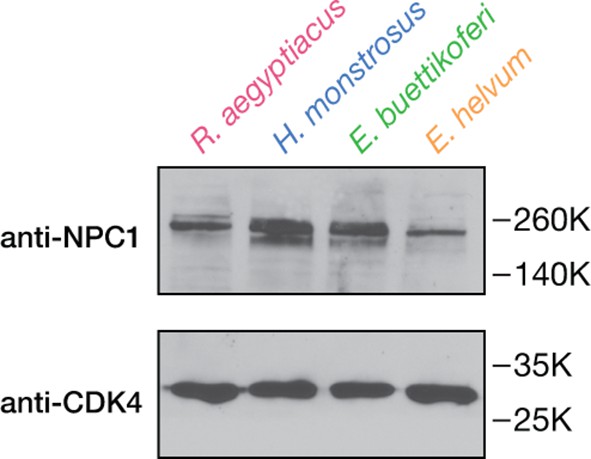
Detection of endogenous NPC1 in pteropodid kidney fibroblast cell lines.
NPC1 in cell extracts was detected by SDS-PAGE and imunoblotting with an anti-NPC1 antibody specific to the NPC1 cytoplasmic tail. Cyclin-dependent kinase 4 (CDK4) was used as a loading control.
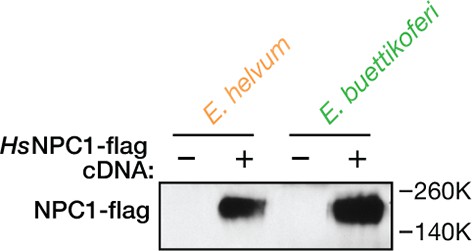
Ectopic expression of human NPC1 in pteropodid kidney fibroblast cell lines.
Cytoplasmic extracts of pteropodid kidney fibroblast cell lines (control or ectopically expressing flag epitope-tagged human NPC1 [HsNPC1]) were resolved in SDS-polyacrylamide gels and detected by immunoblotting with an anti-flag antibody.
The incompatibility between EBOV GP and Eidolon helvum NPC1 reduces, but does not eliminate, EBOV entry into African straw-colored fruit bat cells.
(A) CRISPR/Cas9 genome engineering was used to knock out the NPC1 gene in African straw-colored fruit bat kidney fibroblasts. WT NPC1 gene sequence aligned with the sequences of all three alleles in the knockout (NPC1–#1) cell clone. The gRNA target sequence is marked in red, and the protospacer adjacent motif (PAM) sequence of the gRNA target site is underlined. (B) The capacity of WT and NPC1–#1 cells, and NPC1–#1 cells stably expressing HsNPC1, to clear lysosomal cholesterol was determined by staining with filipin III complex from Streptomyces filipensis, as described (Carette et al., 2011). Red arrowheads indicate lysosomes with accumulated cholesterol. (C) Infection of African straw-colored fruit bat cell lines and Vero African grivet monkey cells (control) by VSVs bearing EBOV or MARV GP. Infected (eGFP-positive) cells were visualized by fluorescence microscopy. Representative fields are shown. Scale bars, 20 µm.
Alignment of bat NPC1 domain C amino acid sequences.
Conserved cysteine residues are labeled in red.Residues under positive selection (see Figure 5, Figure 5—figure supplement 1, and text) are labeled in yellow. Predicted secondary structures for H. sapiens, R. aegyptiacus, and E. helvum domain C were generated using PSIPRED and are shown below the alignment.
African straw-colored fruit bat NPC1 binds poorly to EBOV GP because of a single amino acid change relative to NPC1 from permissive African pteropodids.
(A) Binding of filovirus GP proteins to soluble NPC1 domain C proteins derived from African pteropodids measured by an ELISA. RaNPC1, Egyptian rousette; EbNPC1, Büttikofer’s epauletted fruit bat; HmNPC1, Hammer-headed bat; EhNPC1, African straw-colored fruit bat. (B) A chimera between RaNPC1 and EhNPC1 domain Cs fully rescues EBOV GP-EhNPC1 binding. (C) A single amino acid change in EhNPC1 domain C, F502D, renders it fully competent to recognize EBOV GP. Means ± SD (n = 3) from a representative experiment are shown.
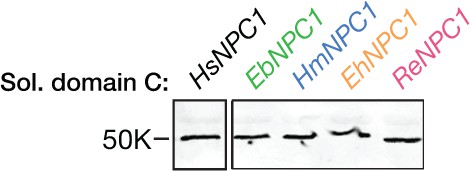
Expression of soluble pteropodid NPC1 domain C proteins.
Flag epitope-tagged NPC1 domain C proteins derived from pteropodids were expressed in 293T cells, subjected to deglycosylation with protein N–glycosidase F, resolved in SDS-polyacrylamide gels, and detected by immunoblotting with an anti-flag antibody.

Amino acid residue 502 is conserved in NPC1 domain C sequences from additional wild-caught African straw-colored fruit bats.
NPC1 sequences were amplified and sequenced from three African straw-colored fruit bat (Eidolon helvum) cell lines and spleen samples derived from nine additional wild-caught E. helvum bats. Alignments of domain C amino acid sequences are shown. Residue 502 is highlighted in orange and white text-on-blue shading. Other amino acid changes are highlighted in green.
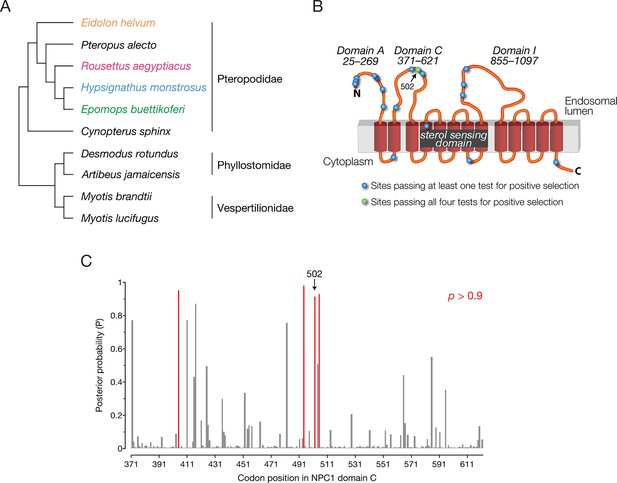
NPC1 is under positive selection in bats.
(A) Bat species included in the evolutionary analysis of NPC1. Family relationships are indicated at the right of the sequence alignment. (B) Positions identified with dN/dS>1 are illustrated on a cartoon schematic of NPC1. Sites in blue were identified in at least one of the four evolutionary analyses performed, and the site in green was identified in all four analyses (Figure 5—figure supplement 1). (C) The posterior probability that each codon in domain C has dN/dS>1 according to PAML. Position 502 is indicated (p = 0.921), and two clusters of sites with elevated posterior probabilities are evident.
Four tests for positive selection in bat NPC1.
The alignment of bat NPC1 orthologs was analyzed with four common tests for positive selection (see Materials and Methods for details). Reported in the tables are the codons that were identified by each test to be experiencing positive selection, as described in the footnotes. Two different sets of analyses were performed, one using the species tree (top) and the other using the gene tree (bottom); both trees are shown and contrasted at the bottom.
A sequence polymorphism in the NPC1-binding site of filovirus GP influences GP-EhNPC1 binding and EhNPC1-dependent filovirus entry.
(A) Binding of EBOV GP (WT and mutant V141A) to soluble NPC1 domain C proteins derived from African pteropodids measured by an ELISA. RaNPC1, Egyptian rousette; EbNPC1, Büttikofer’s epauletted fruit bat; HmNPC1, Hammer-headed fruit bat; EhNPC1, African straw-colored fruit bat. (B) Infection of African straw-colored fruit bat cells with VSV pseudotypes bearing EBOV GP (WT or V141A). Means ± SD (n = 3–4) from a representative experiment are shown in each panel. Means for VSV-EBOV GP WT vs V141A infection were compared by unpaired two-tailed Student’s t-test with Welch’s correction (**p < 0.01). (C) Surface-shaded representation of a single GP1-GP2 monomer (PDB ID: 3CSY (Lee, et al., 2008) highlighting key residues in the NPC1-binding site (yellow) and residue 141 (red). GP1, blue. GP2, grey. (D) Alignments of GP1 sequences from a panel of filoviruses. V141, orange; A141, white text on blue shading; other residues divergent from consensus sequence, black text on green shading. (E) Infection of African pteropodid cells with VSV pseudotypes bearing SUDV GP (WT or A141V). Means ± SD (n = 4) from two biological replicates are shown. Means for VSV-SUDV GP WT vs A141V infection on each cell line were compared by unpaired two-tailed Student’s t-test with Welch’s correction (*p < 0.05, **p < 0.01, ****p < 0.0001).
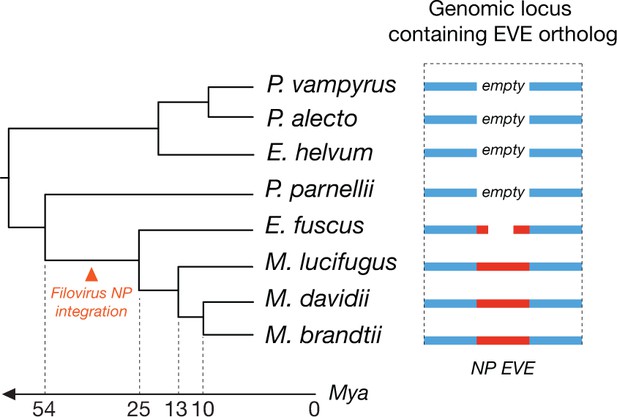
Orthologous endogenous viral elements (EVEs) derived from filovirus nucleoprotein (NP) genes indicate that filoviruses have infected bats for at least 25 million years.
The time-calibrated phylogeny shown to the left is based on estimates obtained in Miller-Butterworth et al., 2007. The schematic to the right shows the orthologous EVEs and empty insertion sites as they occur in each bat genome. Also see Supplementary file 5.
Additional files
-
Supplementary file 1
The sequence of the receptor-binding site in GP is highly conserved among EBOV isolates.
Alignment of GP amino acid sequences corresponding to the NPC1-binding site (residues 53-200) derived from diverse EBOV isolates (listed GenBank accession numbers) is shown. Amino acid changes are highlighted in pink.
- https://doi.org/10.7554/eLife.11785.017
-
Supplementary file 2
Sources of RNAseq data used to assemble NPC1 sequences.
- https://doi.org/10.7554/eLife.11785.018
-
Supplementary file 3
NPC1 sequencing strategy and primers.
- https://doi.org/10.7554/eLife.11785.019
-
Supplementary file 4
NPC1 nucleotide sequences determined experimentally or assembled from publicly available RNAseq data.
- https://doi.org/10.7554/eLife.11785.020
-
Supplementary file 5
Alignment of sequences flanking orthologous filovirus-related EVEs and empty insertion sites.
Multiple sequence alignment showing synteny between sequences flanking filovirus NP-derived EVEs in the indicated bat genomes. (A) The region flanking the 5’ end of the EVE ortholog. (B) Amino acid sequence alignment of a conserved region within exogenous filovirus NP and the putative EVE ORFs found of these sequences (note: the E. helvum sequence in deleted in this region and is not shown). (C) Region flanking the 3’ end of the EVE ortholog. Nucleotide insertions that were present in only one sequence are not shown. EVE sequence is highlighted in grey. Accession numbers for EVE sequences are as follows; M. brandtii (ANKR01230743.1); M. lucifugus (AAPE02014310.1); M. davidii (ALWT01026193.1); E. fuscus (ALEH01076399.1); E. helvum (AWHC01132512.1); P. vampyrus (ABRP02039678.1); P. alecto (ALWS01163349.1); P. parnellii (AWGZ01223755.1).
- https://doi.org/10.7554/eLife.11785.021





