Sleep-active neuron specification and sleep induction require FLP-11 neuropeptides to systemically induce sleep
Figures
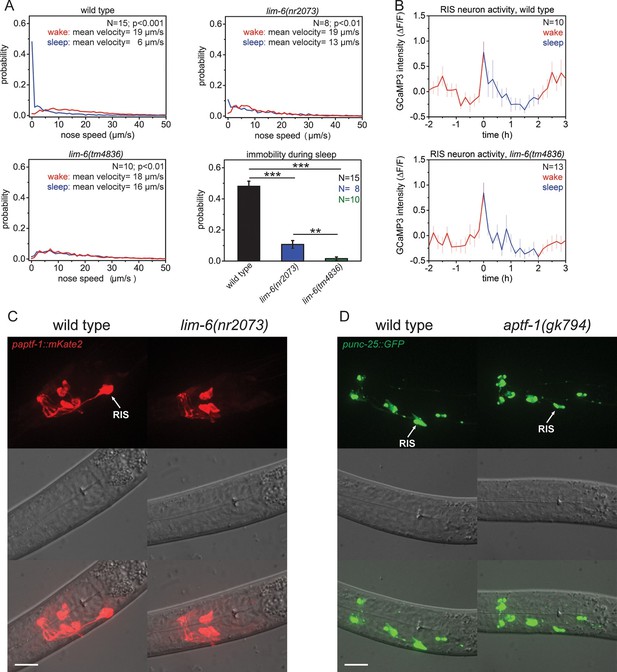
The LIM homeobox transcription factor LIM-6 controls sleep by specifying expression of the transcription factor APTF-1 in RIS.
(A) Probability distribution of nose speeds during wake and sleep for wild type and lim-6 mutants. lim-6(nr2073) shows substantially reduced and lim-6(tm4836) shows a complete lack of immobility during the time the animals should be sleeping. (B) Averaged RIS calcium activity pattern across time in wild type and lim-6(tm4836). RIS is active at the onset of sleep in wild type and in lim-6(tm4836). There was no statistically significant difference between wild-type and lim-6 worms (p > 0.05, Welch test). (C) Expression of paptf-1::mKate2 in wild type and lim-6(nr2073) L1 larvae. Expression of mKate2 is absent in RIS in lim-6(nr2073) showing that LIM-6 controls expression of APTF-1 in RIS. (D) Expression of punc-25::GFP in wild type and aptf-1(gk794). Reporter GFP expression is normal in RIS in aptf-1 mutant worms indicating that GABAergic function is not controlled by APTF-1. Statistical tests used were Wilcoxon Signed Paired Ranks test for comparison within the same genotype and Student’s t-test for comparisons between genotypes. Error bars are SEM. ** denotes statistical significance with p<0.01, *** denotes statistical significance with p<0.001. Scale bars are 10 µm.
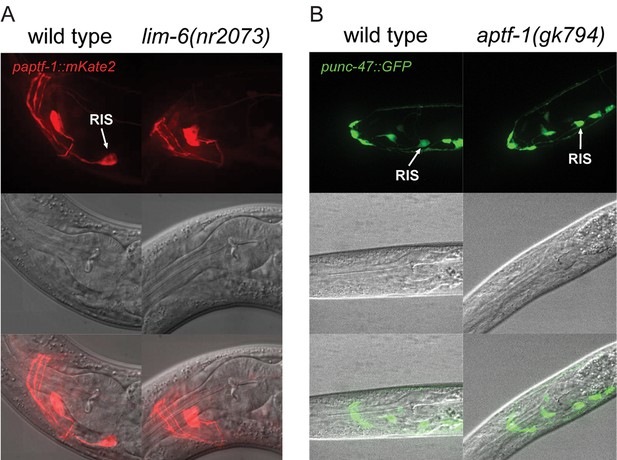
LIM-6 controls expression of APTF-1 across development but does not control the expression of the GABA vesicular transporter gene unc-47.
(A) Expression of paptf-1::mKate2 in wild type and in lim-6(nr2073) L4 larvae. Expression of mKate2 is absent in RIS in lim-6(nr2073) at the L4 stage showing that LIM-6 generally controls expression of APTF-1 in RIS rather than the onset of expression. Scale bar is 10 µm. (B) punc-47::GFP is expressed in aptf-1(gk794) mutant worms. Shown are L1 larvae. Scale bar is 10 µm.
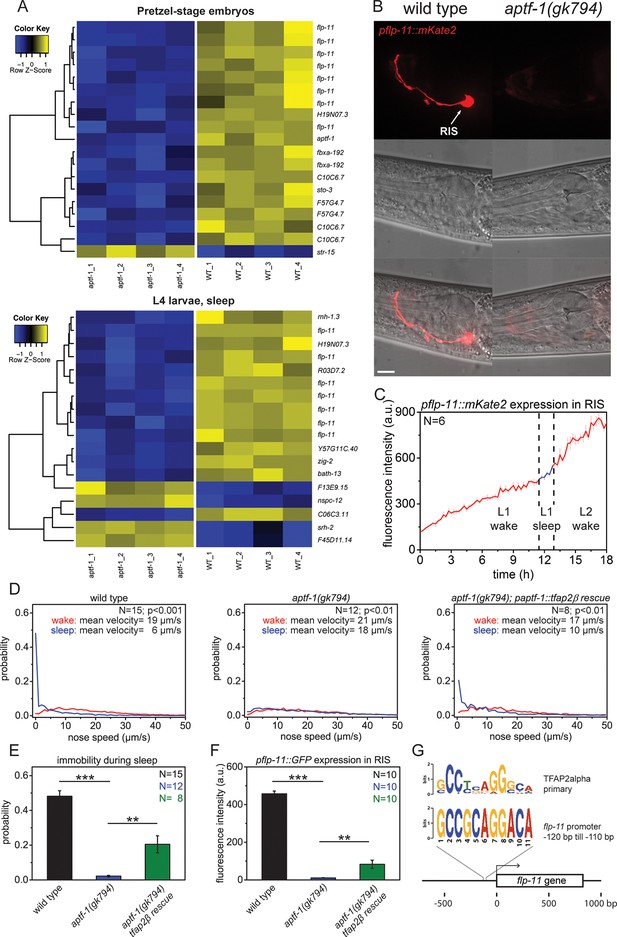
The AP2 transcription factor APTF-1 controls FLP-11 expression in RIS.
(A) Transcriptional analysis of aptf-1(gk794) mutants revealed genes that are regulated by APTF-1. Wild-type and aptf-1(gk794) pretzel-stage embryos and sleeping L4 larvae were used for a transcriptome analysis. In both life stages, expression of the FMRFamide-like neuropeptide FLP-11 was strongly reduced in aptf-1(gk794). This suggests transcriptional control of FLP-11 by APTF-1. Data can be found in Supplementary file 1, Tables 1A and 1B. (B) Expression of pflp-11::mKate2 in wild type and aptf-1(gk794). Expression of mKate2 was absent in RIS in aptf-1(gk794) showing that APTF-1 controls expression of FLP-11. Expression of flp-11 in RIS was reminiscent to the expression of the flp-11 homolog afp-6 in RIS in Ascaris nematodes (Yew et al., 2007). Expression for additional genes can be found in Figure 2—figure supplements 2 and 3. (C) flp-11 expression profile in RIS over the sleep-wake cycle. Expression does not change with the sleep-wake cycle. (D) Probability distribution of nose speeds during wake and sleep for wild type, aptf-1(gk794) and aptf-1(gk794); paptf-1::tfap2β rescue. (E) Comparison of immobility during sleep for wild type, aptf-1(gk794), and aptf-1(gk794); paptf-1::tfap2β. The mouse TFAP2β partially rescued the aptf-1(gk794) sleep phenotype. (F) Comparison of pflp-11::GFP fluorescence intensity in RIS for wild type, aptf-1(gk794), and aptf-1(gk794); paptf-1::tfap2β. The mouse TFAP2β partially rescued the expression of flp-11 in RIS (18% of wild-type level). (G) Analysis of putative AP2-binding sites in the flp-11 promoter region. The flp-11 promoter region was scanned for the primary mouse AP2α-binding site. Overlap was found (p<0.001, q=0.06 (Grant et al., 2011)) for one binding site. Statistical test used was Wilcoxon Signed Paired Ranks test. ** denotes statistical significance with p<0.01, *** denotes statistical significance with p<0.001. Scale bar is 10 µm.
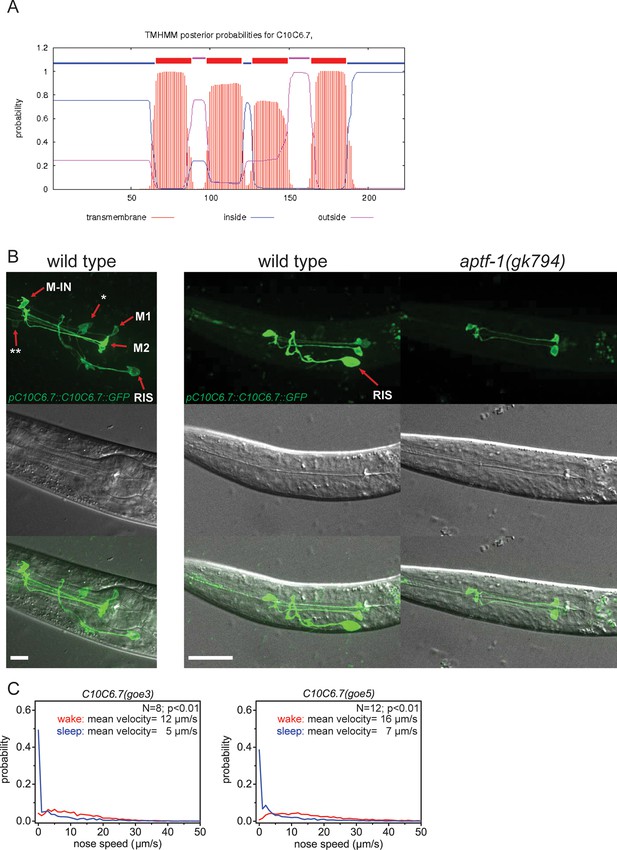
C10C6.7 is a putative four transmembrane helix protein that is expressed in RIS and that is controlled by aptf-1.
(A) Bioinformatics analysis suggests that C10C6.7 is a four transmembrane helix protein (Krogh et al., 2001). (B) Expression pattern of GFP-tagged fosmids for C10C6.7. Expression is visible in nine cells: interneuron RIS; pharyngeal neurons M1, M2, Motor-interneuron (M-IN), an unidentified pair of pharyngeal neurons (**) and an unidentified pair of sensory neurons (*). Expression of C10C6.7 protein in RIS is controlled by aptf-1. (C) Probability distribution of nose speeds during wake and sleep for C10C6.7(goe3) and C10C6.7(goe5) shows that C10C6.7 does not play a significant role in sleep control. Statistical test used was Wilcoxon Signed Paired Ranks test. Scale bars are 10 µm.
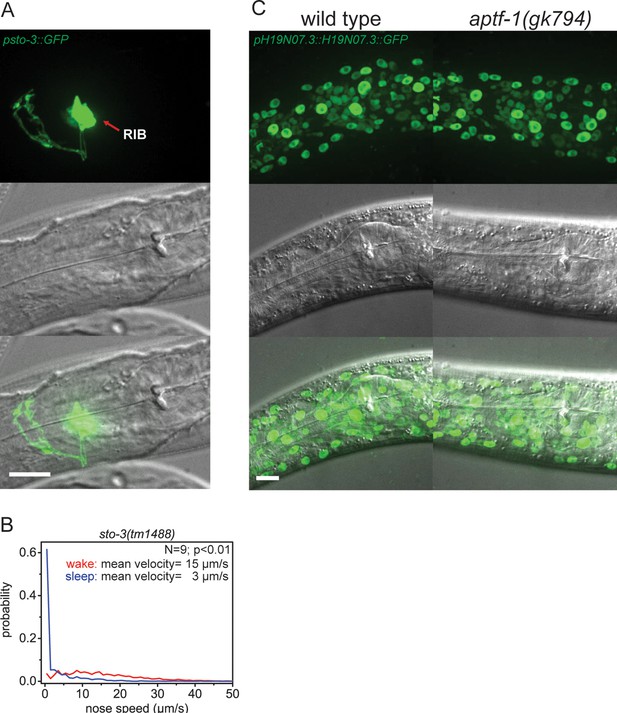
Expression pattern of sto-3 and H19N07.3.
(A) Expression pattern of sto-3 promoter fusions. STO-3 is expressed in RIB neuron and additionally in three unidentified non-neuronal cells in the tale (not shown). (B) Probability distribution of nose speeds during wake and sleep for sto-3(tm1488) shows that sto-3 does not play a significant role in sleep control. (C) Expression pattern of GFP-tagged fosmids for H19N07.3. The H19N07.3 protein is expressed in all somatic cell nuclei, but its levels are not regulated by aptf-1. Statistical test used was Wilcoxon Signed Paired Ranks test. Scale bars are 10 µm.
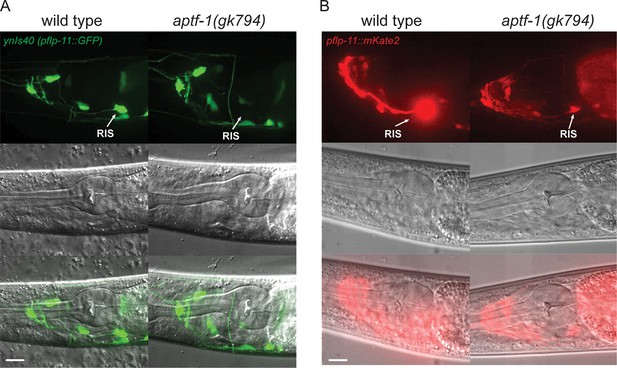
FLP-11 is strongly expressed in RIS and weakly in additional neurons. APTF-1 controls the expression in RIS.
(A) Expression pattern of ynIs40(pflp-11::GFP) in wild type and aptf-1(gk794) mutant. The transgene expresses in several neurons including RIS. aptf-1(gk794) abolishes the expression specifically in RIS. (B) Expression pattern of goeIs288(pflp-11::mKate2) in wild type and the aptf-1(gk794) mutant. Strong expression is visible only in RIS. By increasing the contrast to the point where RIS is over-saturated several additional neurons becomes visible that may be identical to those seen in ynIs40 (Kim and Li, 2004). aptf-1(gk794) strongly reduces the expression specifically in RIS. Scale bars are 10 µm.
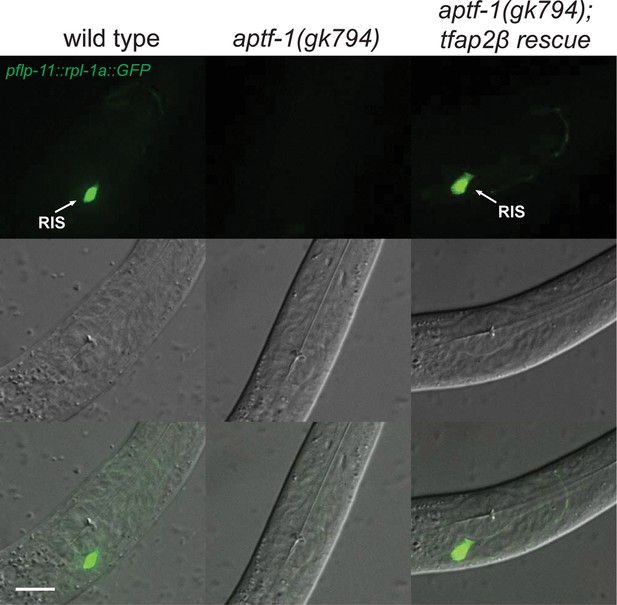
Mouse TFAP2beta partially restores expression of flp-11 neuropeptides in RIS in aptf-1 mutant worms.
Expression of pflp-11::GFP in wild type, aptf-1(gk794), and aptf-1(gk794); tfap2beta rescue. In aptf-1(gk794), expression of flp-11 is strongly reduced but could partially be restored by the mouse TFAP2beta. Expression of flp-11::GFP in the rescue strain varied between 5–50% of wild-type levels. Here, we show a picture of 50% rescue. Scale bar is 10 µm.
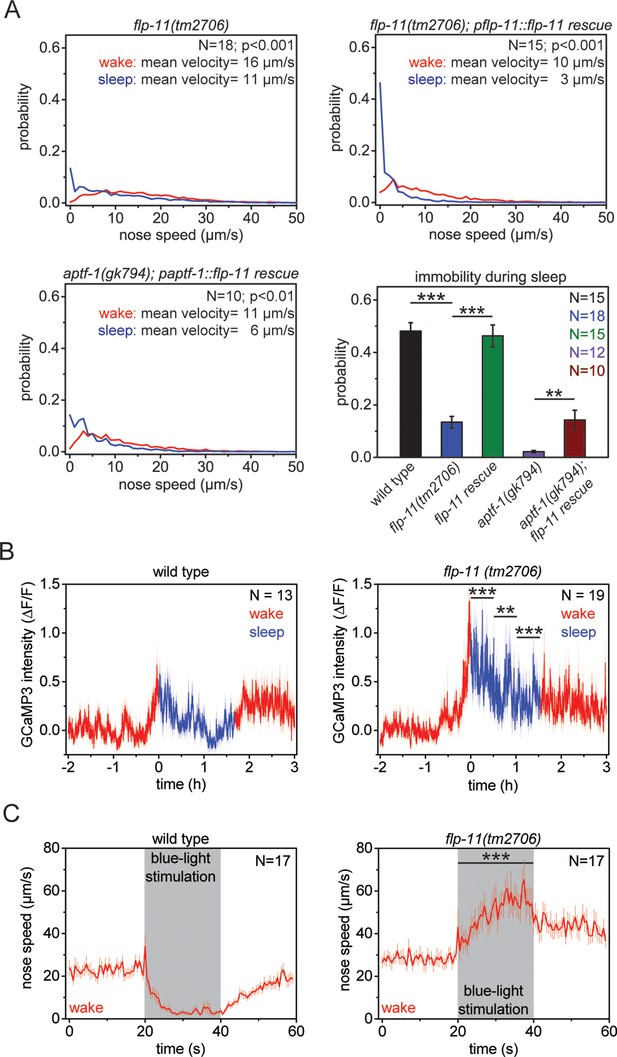
RIS induces sleep through the sleep-inducing FMRFamide-like neuropeptide FLP-11.
(A) Probability distribution of nose speeds during wake and sleep for wild type, flp-11(tm2706), flp-11(tm2706); pflp-11::flp-11 rescue and aptf-1(gk794); paptf-1::flp-11 rescue. Immobility during the time the animal should be sleeping was substantially reduced in flp-11(tm2706). flp-11(tm2706) could be rescued by expression of the wild-type flp-11 gene. Furthermore, expression of flp-11 in aptf-1(gk794) partially rescued sleep behavior. (B) Averaged RIS calcium activity pattern across time in wild type and flp-11(tm2706). RIS was strongly activated at the onset of sleep in flp-11(tm2706) (Student’s t-test). (C) Channelrhodopsin-2 activation of aptf-1-expressing neurons caused immediate immobility in wild type. In contrast, flp-11(tm2706) accelerated upon blue light stimulation showing that RIS-dependent immobility is impaired. Statistical tests used were Wilcoxon Signed Paired Ranks test for comparisons within genotypes and Student’s t-test for comparisons between genotypes. Error bars are SEM. ** denotes statistical significance with p<0.01, *** denotes statistical significance with p<0.001.
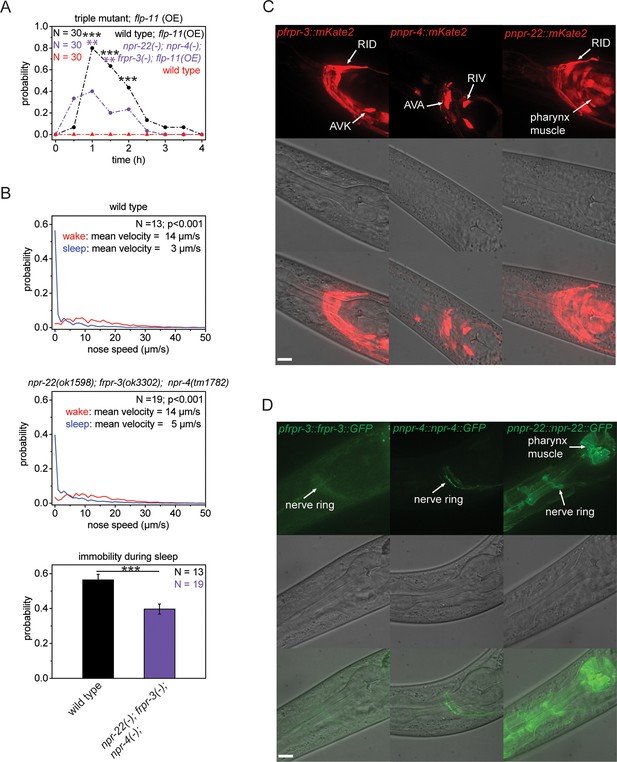
Multiple receptors may be involved in sleep induction.
(A) Behavioral analysis over time after the heat-shock-induced overexpression of flp-11 in wild-type and the npr-22(ok1598); frpr-3(ok3302); npr-4(tm1782) triple mutant. To assess the effect of the heat shock on quiescence, wild-type worms without flp-11 overexpression were analyzed at the same time and do not show any behavioral changes. Overexpression of flp-11 caused anachronistic quiescence that was lasting approximately 1 hr in the wild type. Quiescence was significantly reduced by approximately 50% in the triple mutant. (B) Probability distribution of nose speeds during wake and sleep for wild type and npr-22(ok1598); frpr-3(ok3302); npr-4(tm1782) triple mutant. Immobility during sleep was reduced by about 30% in the npr-22(ok1598); frpr-3(ok3302); npr-4(tm1782) triple mutant. (C) Expression patterns of frpr-3, npr-4 and npr-22 promoter fusions. FRPR-3 is expressed in about 30 neurons, mostly in the head. Expression of NPR-4 was seen in about five neurons. NPR-22 was expressed in several neurons and muscle tissue including pharynx and head muscle. (D) Expression patterns of GFP-tagged fosmids for frpr-3, npr-4, and npr-22. FRPR-3 and NPR-4 were mostly expressed around the nerve ring. NPR-22 localized broadly to the plasma membrane in several neurons, pharynx muscle, head muscle, and the anal sphincter muscle. Statistical tests used were Wilcoxon Signed Paired Ranks test for comparisons within genotypes and Student’s t-test for comparisons between genotypes. Error bars are SEM. ** denotes statistical significance with p<0.01, *** denotes statistical significance with p<0.001. Scale bars are 10 µm.
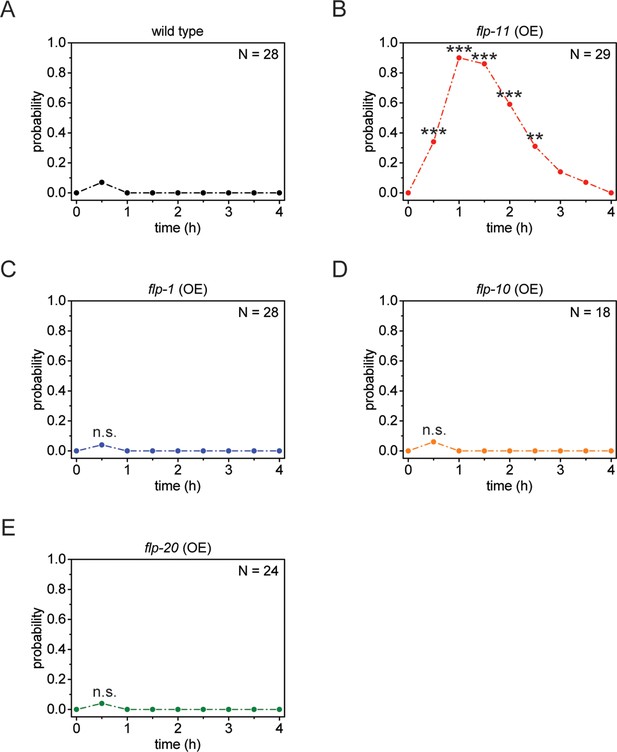
Heat-shock-induced flp-11 overexpression causes quiescence but heat-shock-induced overexpression of three other flp genes does not, suggesting that quiescence cannot be induced by overexpression of any flp.
(A) Behavioral analysis of wild-type adult worms over time after 5 min of heat shock at 37°C. (B-E) Behavioral analysis of wild-type adult worms over time after heat shock-induced overexpression of flp-11, flp-1, flp-10, and flp-20. Only heat-shock-induced overexpression of flp-11 induces quiescence.
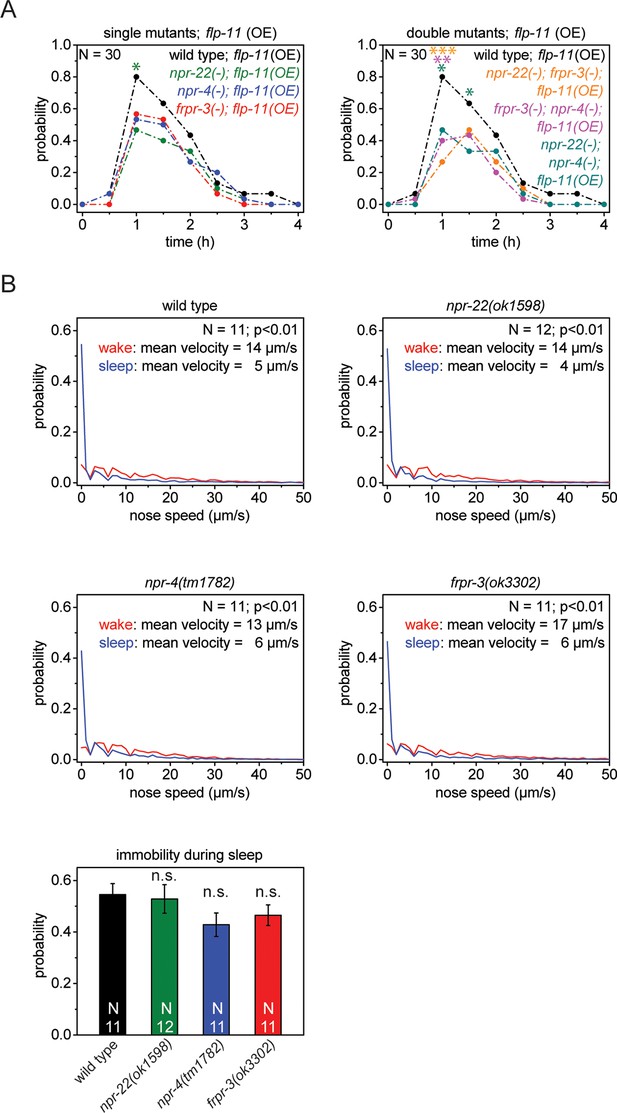
Single receptors mutants do not show reduced quiescence during sleep, but do show reduced quiescence upon heat-shock-induced overexpression of flp-11.
(A) Heat shock-induced overexpression of flp-11 in single and double mutants of frpr-3(ok3302), npr-4(tm1782), and npr-22(ok1598). The strength of the anachronistic quiescence correlates with the combination of receptors ranging from highest for single mutants to lowest for the double mutants. (B) Probability distribution of nose speeds during wake and sleep for wild type and npr-22(ok1598), frpr-3(ok3302), npr-4(tm1782) single mutants. Immobility during sleep is not significantly different between wild type and single mutants. Statistical tests used were Wilcoxon Signed Paired Ranks test for comparisons within genotypes and Student’s t-test for comparisons between genotypes. Error bars are SEM. *denotes statistical significance with p<0.05, **denotes statistical significance with p<0.01, *** denotes statistical significance with p<0.001.
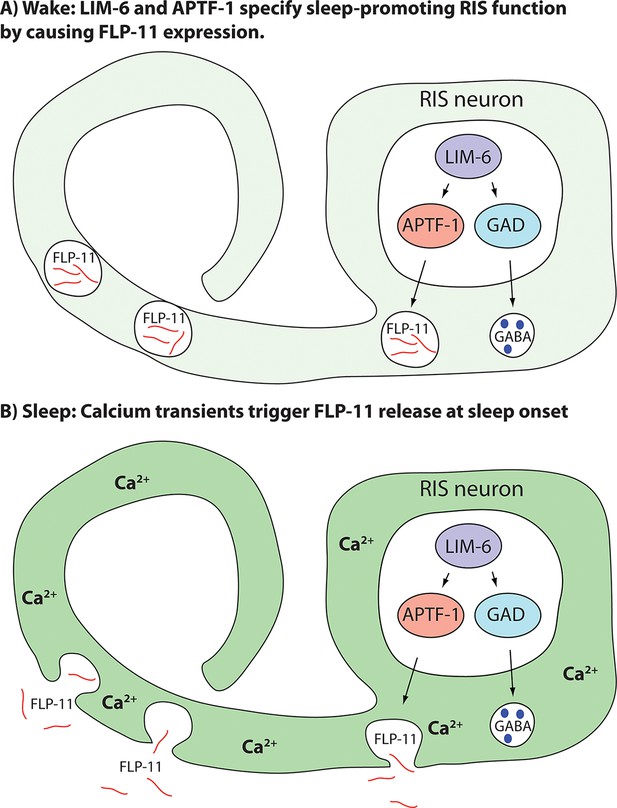
Model for generation of sleep-promoting function of RIS and sleep induction by RIS.
According to this model, the transcription factor LIM-6 controls GABAergic and peptidergic function in RIS in parallel. To render this neuron sleep-promoting, LIM-6 is required for the expression of the APTF-1 transcription factor. APTF-1, in turn, is required for the expression of sleep-inducing FLP-11 peptides. FLP-11 is present in RIS at all times. Sleep onset is triggered by an unknown signal, which leads to a depolarization and to calcium influx. This triggers FLP-11 release, which in turn systemically induces sleep behavior.
Additional files
-
Supplementary file 1
Genes that are differentially expressed in aptf-1(-).
Table 1A, Genes with altered expression in aptf-1(gk794) mutant in pretzel-stage embryos. Table 1B, Genes with altered expression in aptf-1(gk794) mutant during L4 larvae sleep.
- https://doi.org/10.7554/eLife.12499.015
-
Supplementary file 2
C. elegans strains.
A list of C. elegans strains that were used for this study.
- https://doi.org/10.7554/eLife.12499.016
-
Supplementary file 3
Primers.
A list of primers that were used for this study.
- https://doi.org/10.7554/eLife.12499.017
-
Supplementary file 4
DNA constructs.
A list of DNA constructs (Plasmids and Fosmids) that were used for this study.
- https://doi.org/10.7554/eLife.12499.018





