Insight into the evolution of microbial metabolism from the deep-branching bacterium, Thermovibrio ammonificans
Figures
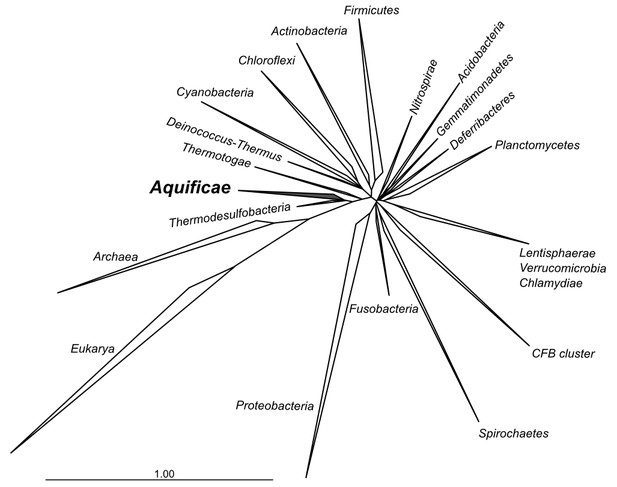
Phylogenetic tree of 16S rRNA sequences computed from the current version of the aligned 16S rRNA database obtained from the arb-SILVA project (http://www.arb-silva.de/).
https://doi.org/10.7554/eLife.18990.003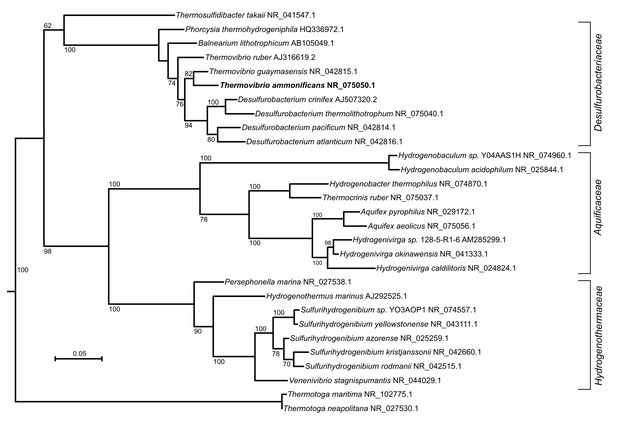
Maximum likelihood tree showing the 16S rRNA relationship of the Aquificae phylum.
Thermotoga spp. were used as outgroup. Bootstrap values based on 1000 replication are shown at branch node. Bar, 5% estimated substitution.
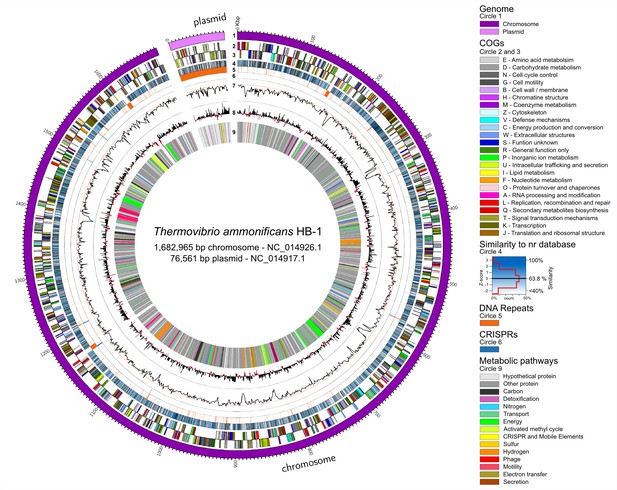
Thermovibrio ammonificans HB-1 genomic map highlighting main genomic features.
The circular chromosome and plasmid are drawn together. Several other repeats were identified in the chromosome (circle 5). From the outer circle: 1 – Dimension of the genome in base pairs (chromosome in violet, plasmid in pink); 2 and 3 – sense and antisense coding sequences colored according to their COGs classification (see legend in the figure); 4 – similarity of each coding sequence with sequences in the non-redundant database; 5 – position of tandem repeat in the genome. A total of 46 tRNA were identified, comprising all of the basic 20 amino acid plus selenocysteine; 6 – position of CRISPRs; 7 – GC mol% content, the red line is the GC mean of the genome (52.1 mol%); 8 – GC skew calculated as G+C/A+T; 9 – localization of coding genes arbitrarily colored according to the metabolic pathways reconstructed (see Figure 1). Colors are consistent trough the entire paper. Total dimension and accession number for the chromosome and plasmid are given.
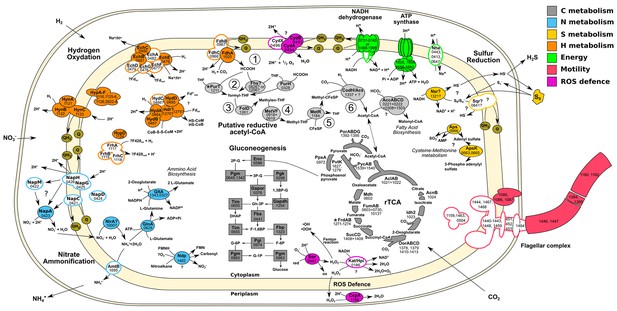
Central metabolism of T. ammonificans HB-1.
Enzyme names are reported together with the gene locus number (Theam_number). Primary compounds involved in reactions were also reported, however visualized reactions are not complete. Pathways were arbitrarily color coded according to their reconstructed function and are consistent throughout the paper. Solid shapes represent genes for which the enzyme was found in the proteome, while outlined shapes were only identified in the genome. Circled numbers 1 to 6 represent reaction numbers of the putative reductive acetyl-CoA pathway as described in the text. Abbreviations: NITRATE AMMONIFICATION: NapCMADGH – Periplasmic nitrate reductase complex; NirA – Putative nitrite reductase; AmtB – ammonia transporter; GlnA – L-Glutamine synthetase; GltA – Glutamate synthetase. HYDROGEN OXIDATION: HynABC – Ni-Fe Membrane bound hydrogenase; FrhACB – Cytosolic Ni-Fe hydrogenase/putative coenzyme F420 hydrogenase; HupD – Cyrosolic Ni-Fe hydrogenase maturation protease; HypA-F – Hydrogenases expression/synthesis accessory proteins; EchABCEF – Ech membrane bound hydrogenase complex; HydAB – Cytosolic Ni-Fe hydrogenases potentially involved in ferredoxin reduction; Hdr? – Missing heterodisulfide reductase CoB-CoM; FdhABC – Formate dehydrogenase. ENERGY PRODUCTION: NADH dehydrogenase and ATP synthetase are reported without the names of the single units; Nhe – Sodium/hydrogen symporter. SULFUR REDUCTION: Sqr – Putative sulfate quinone reductase involved in sulfur respiration; Nsr? – FAD/NAD nucleotide-disulphide oxidoreductase; Aps – Sulfate adenylyl transferase and kinase involved in assimilation of sulfur. FLAGELLAR COMPLEX: for simplicity single unit names are not reported. REDUCTIVE ACETYL-CoA: Fhs – Proposed reverse formyl-THF deformylase; PurHT – Formyltransferases (phosphoribosylaminoimidazolecarboxamide and phosphoribosylglycinamide) FolD – Methenyl-THF cyclohydrolase and dehydrogenase; MetVF – Methylene-THF reductase; MetR – Putative methyl-transferase; CodH – Carbon monoxyde dehydrogenase; AcsA – Acetyl-CoA ligase/synthase; AccABCD – Acetyl-CoA carboxylase. REDUCTIVE CITRIC ACID CYCLE: AclAB – ATP-citrate lyase; Mdh – Malate dehydrogenase; FumAB – Fumarate hydratase; FrdAB – Fumarate reductase; SucCD – Succinyl-CoA synthetase; OorABCD – 2-Oxoglutarate synthase; Idh2 – Isocitrate dehydrogenase/2-oxoglutarate carboxylase; AcnB – Aconitate hydratase; PorABDG – Pyruvate synthase; PycAB - Pyruvate carboxylase; PpsA – Phosphoenolpyruvate synthase water dikinase; PyK – Pyruvate:water dikinase. GLUCONEOGENESIS: Eno – Enolase; Pgm – Phosphoglycerate mutase; Pgk – Phosphoglycerate kinase; Gapor – Glyceraldehyde-3-phosphate dehydrogenase; Gapdh – Glyceraldehyde 3-phosphate dehydrogenase; Fba – Predicted fructose-bisphosphate aldolase; Tim – Triosephosphate isomerase; Fbp – Fructose-1,6-bisphosphatase I; Pgi – Phosphoglucose isomerase; Pgm – Phosphoglucomutase.
-
Figure 3—source data 1
List of the proteins identified in the proteome of T. ammonificans grown under nitrate reducing conditions.
- https://doi.org/10.7554/eLife.18990.008
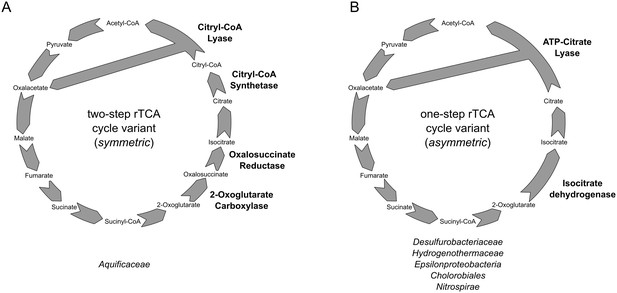
Reductive TCA cycle variants found in extant bacterial lineages.
(A) two-step variant of the rTCA cycle, also known as symmetric variants, were the carboxylation of 2-oxoglutarate is performed in two steps by the enzymes 2-oxoglutarate carboxylase and oxalosuccinate reductase, and the cleavage of citrate is performed in two step by the citryl-CoA synthetase and citryl-CoA lyase enzymes; (B) one-step variant of the rTCA cycle, also known as asymmetric variants, were the carboxylation of 2-oxoglutarate is performed in a single step by the enzymes isocitrate dehydrogenase, and the cleavage of citrate to acetyl-CoA is performed in a single reaction by the ATP citrate lyase enzyme.
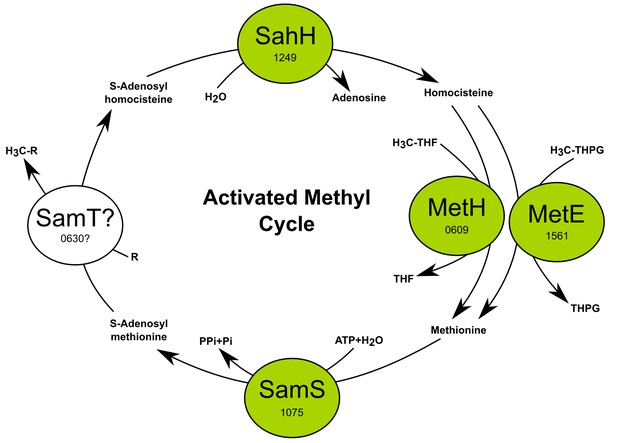
The activated methyl cycle of T. ammonificans reconstructed from the genome.
https://doi.org/10.7554/eLife.18990.010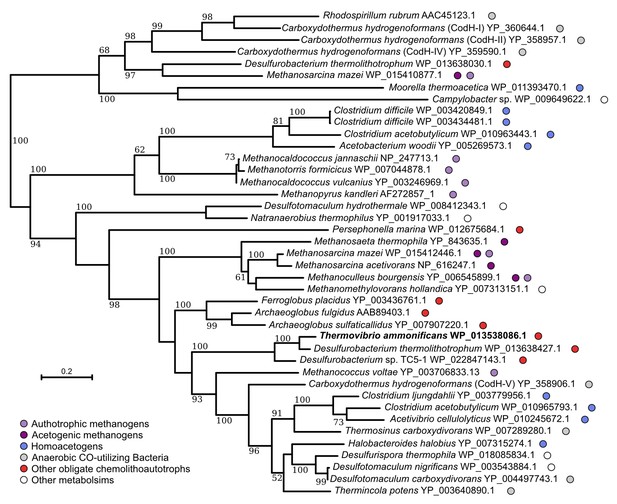
Neighbor-joining tree showing the position of the carbon monoxide dehydrogenase, CodH, of T. ammonificans (in bold).
Bootstrap values based on 1000 replications are shown at branch nodes. Only bootstrap values above 50% are reported. Bar, 10% estimated substitutions. The metabolism of each organism is reported on the side.
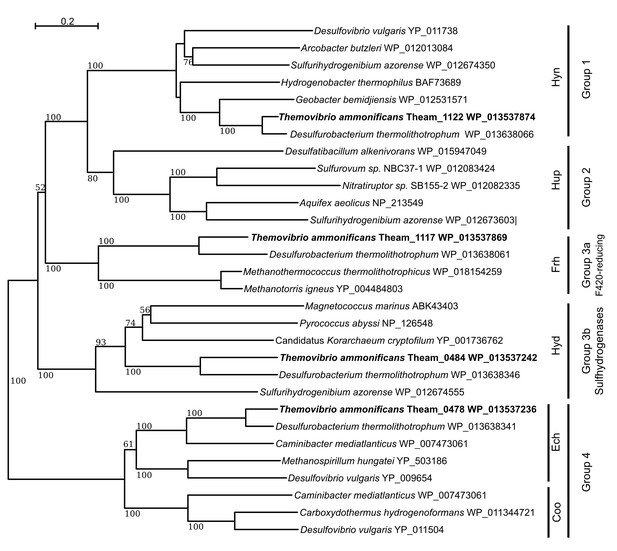
Neighbor-Joining tree showing the position and classification of the [NiFe]-hydrogenases found in the genome of T.ammonificans.
Bootstrap values based on 1000 replication are shown at branch node. Bar, 20% estimated substitution.
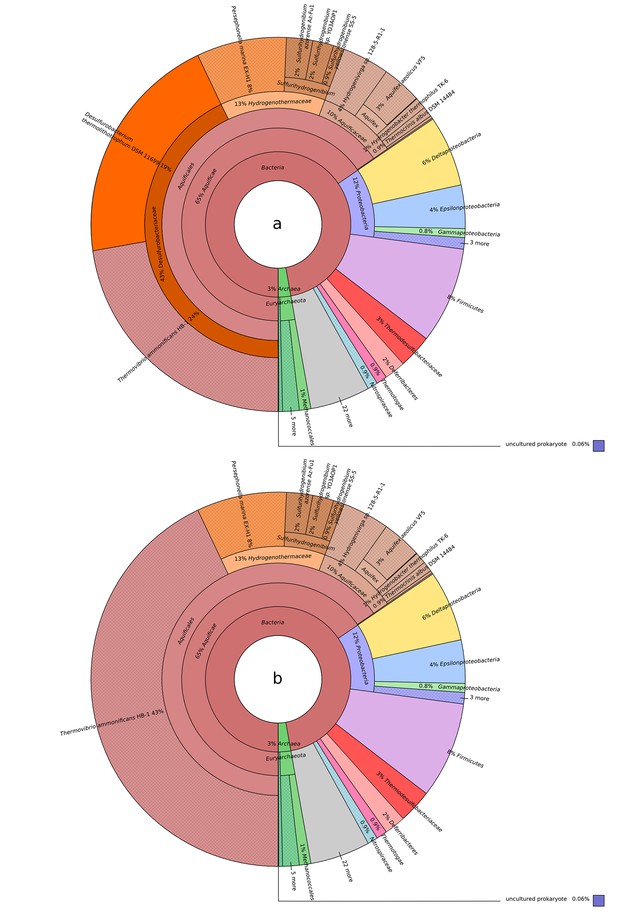
Blastp best hit (cut off at 40% similarity) for the T. ammonificans CDS: (a) Desulfurobacterium thermolithotrophum is included in the database; (b) D. thermolithotrophum and Desulfurobacterium sp.
TC5-1 are excluded from the database (best hits are outside of the Desulfurobacteriaceae family). The interactive versions of the Krona plots are available for download at DOI: 10.6084/m9.figshare.3178528.
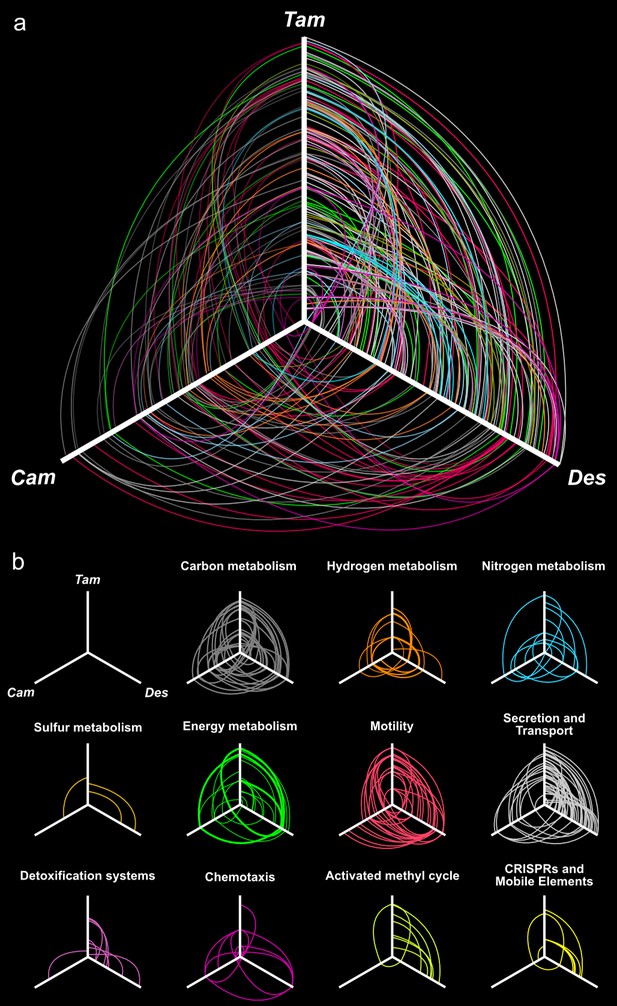
Hives plot presenting the comparative genomic analysis of the T. ammonificans (Tam) genome with the closest relative available genomes of D. thermolithotrophum (Des) and the ecologically similar Epsilonproteobacterium C. mediatlanticus (Cam).
The three axes represent the organism’s linearized genomes with the origin of replication at the center of the figure. Genome size was normalized for visualization purpose. (A) Localization of homologous genes and syntenic regions among the three genomes. Lines colored according to the general pathway code adopted in Figure 3 connect homologous coding sequences. Lines opacity is proportional to similarity between sequences (darker = higher similarity), line width is proportional to the extent of the syntenic area. (B) Hives panel of the homologous genes divided by pathways. Collinear lastZ alignments and interactive dot-plot replicating the analysis are accessible at the GenomeEvolution website with the following permanent addresses: T. ammonificans and D. thermolithotrophum (http://genomevolution.org/r/9vb3); T. ammonificans and C. mediatlanticus (http://genomevolution.org/r/9vb4); D. thermolithotrophum and C. mediatlanticus (http://genomevolution.org/r/9vb8).
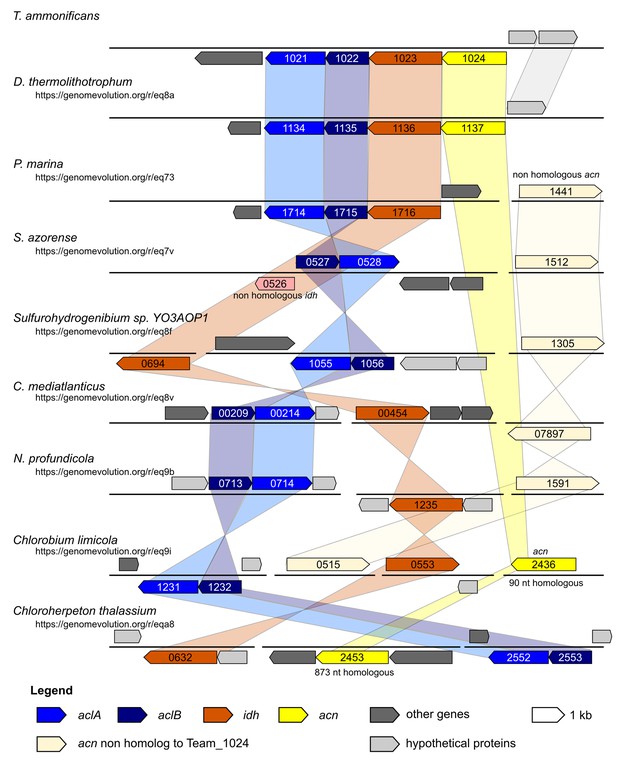
Syntheny diagram presenting the genome organization around the rTCA key enzyme ATP citrate lyase.
In T. ammonificans the two subunits of the ATP citrate lyase enzyme are organized in a single operon together with the isocitrate dehydrogenase and the aconitate dehydratase. The numbers inside each gene represent the locus number for the organism. Shaded color connects synthenic regions. The website address below each organism name is a permanent link to the pairwise analysis performed on the Genome Evolution server (http://genomevolution.org/) against T. ammonificans.
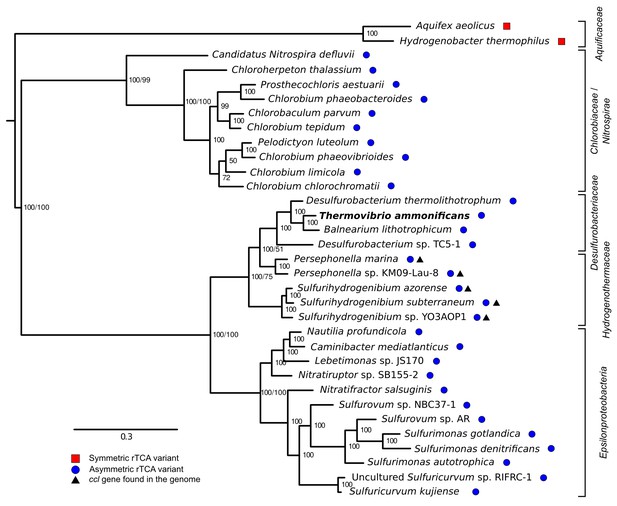
Phylogenetic tree of ATP citrate lyase amino acid sequences.
The tree was constructed with Bayesian inference and maximum likelihood methods and presents the phylogenetic relationship among the concatenated subunit of the ATP citrate lyase in different organisms known to use the rTCA cycle. The numbers near the nodes represent the bayesian posterior probability (left number) and the maximum-likelihood confidence values based on 1000 bootstrap replications (right number). Bar, 30% estimated substitutions. Accession numbers are reported in Figure 9—source data 1. Squares – symmetric rTCA cycle variant characterized by the presence of two-step enzyme reactions for the cleavage of citrate and the carboxylation of 2-oxoglutarate. Circles – asymmetric rTCA cycle variant characterized by one-step reaction enzymes catalyzing the above reactions. Triangles – presence of a citryl-CoA lyase in the genome. See Supplementary Materials for details.
-
Figure 9—source data 1
Accession number of the ATP citrate lyase sequences used for the reconstruction of the phylogenetic history of the enzyme presented in Figure 2 in the main text.
- https://doi.org/10.7554/eLife.18990.019
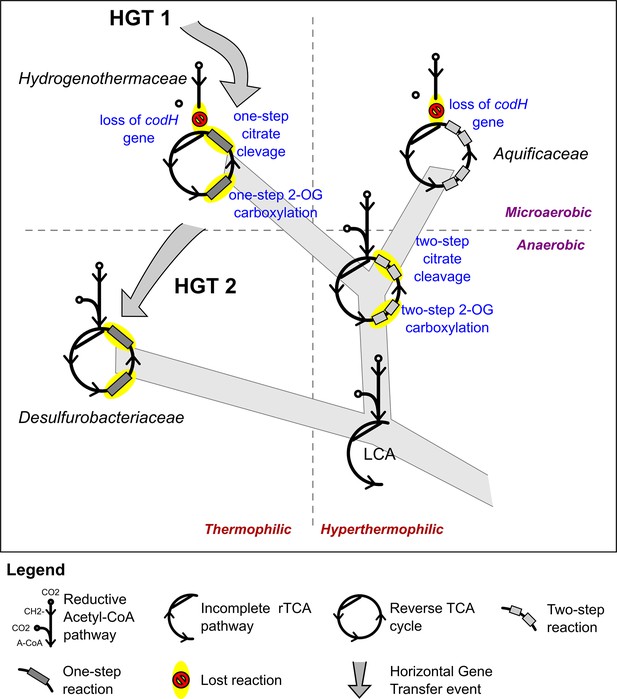
Proposed evolution of the carbon fixation pathway in the Aquificae phylum.
Proposed evolution of the carbon fixation pathway in the Aquificae phylum and reconstruction of the ancestral carbon fixation for the last common ancestor (LCA) based on the results of integrated phylogenetic, comparative genomic and phylometabolic analyses.
Tables
Characteristic of representative members of Aquificae phylum.
| Family | Organism | Growth temp | Energy source | Carbon source | Terminal electron acceptor | Isolated from | Genome sequence accession number | References |
|---|---|---|---|---|---|---|---|---|
| Aquificaceae | Aquifex aeolicus VF5 | 95°C | H2 | CO2 | O2 | Underwater volcanic vents, Aeolic Islands Sicily, Italy | NC_000918.1 | (Deckert et al., 1998; Huber et al., 1992) |
| Hydrogenivirga sp.128–5 R1-11 | 75°C | S2O32-, S0 | CO2 | NO3-, O2 | Lau Basin hydrothermal vent area, Pacific Ocean | NZ_ABHJ00000000 | (Nakagawa et al., 2004; Reysenbach et al., 2009) | |
| Hydrogenobacter thermophilus TK-6 | 75°C | H2 | CO2 | O2 | Hot springs in Izu and Kyushu, Japan | NC_017161.1 | (Arai et al., 2010; Kawasumi et al., 1984) | |
| Hydrogenobaculum sp. Y04AAS1* | - | - | - | - | Marine hydrothermal area, Vulcano Island, Italy | NC_011126.1 | (Reysenbach et al., 2009; Stohr et al., 2001) | |
| Thermocrinis ruber DSM 12173 | 85°C | H2, S2O32-, S0 | CO2 | O2 | Octopus Spring, Yellowstone National Park, Wyoming, USA | PRJNA75073 | (Huber et al., 1998) | |
| Desulfurobacteriaceae | Balnearium lithotrophicum 17S | 70–75˚C | H2 | CO2 | S0 | Deep-sea hydrothermal vent chimney, Suiyo Seamount, Japan | Not sequenced | (Takai et al., 2003b) |
| Desulfurobacterium thermolithotrophum DSM 11699 | 70°C | H2 | CO2 | S0, S2O32-, SO2- | Deep-sea hydrothermal chimney, mid-Atlantic ridge, Atlantic Ocean | NC_015185.1 | (Göker et al., 2011; L'Haridon et al., 1998) | |
| Phorcysia thermohydrogeniphila HB-8 | 75°C | H2 | CO2 | S0, NO3- | Tube of Alvinella pompejana tubeworms, deep-sea hydrothermal vents 13 ˚N, East Pacific Rise, Pacific Ocean | Not sequenced | (Pérez-Rodríguez et al., 2012) | |
| Thermovibrio ammonificans HB-1 | 75°C | H2 | CO2 | S0, NO3- | Deep sea hydrothermal vents 9˚N, East Pacific Rise, Pacific Ocean | NC_014926.1 | (Giovannelli et al., 2012; Vetriani et al., 2004) | |
| Hydrogenothermaceae | Hydrogenothermus marinus VM1 | 65°C | H2 | CO2 | O2(1–2%) | Deep sea hydrothermal vents 9˚N, East Pacific Rise, Pacific Ocean | Not sequenced | (Stohr et al., 2001) |
| Persephonella marina EX-H1 | 73°C | S0 | CO2 | O2, NO3- | Deep sea hydrothermal vents 9˚N, East Pacific Rise, Pacific Ocean | NC_012440.1 | (Götz et al., 2002; Reysenbach et al., 2009) | |
| Sulfurihydrogenibium sp. YO3AOP11 | 70°C | S2O32-, S0 | CO2 | O2 | Calcite Hot Springs, Yellowstone National Park, USA | NC_010730.1 | (Reysenbach et al., 2009; Takai et al., 2003a) | |
| Venenivibrio stagnispumantis | 70°C | H2 | CO2 | O2 | Terrestrial hot spring Champagne Pool, Waiotapu, New Zealand | Not sequenced | (Hetzer et al., 2008) | |
| Incertae sedis | Thermosulfidibacter takai ABI70S6T | 70°C | H2 | CO2 | S0 | deep-sea hydrothermal field at Southern Okinawa Trough, Japan | Not sequenced | (Nunoura et al., 2008) |
-
*– Strain not formally described whose genome sequence is available. For these strains, the physiological information reported in Table S1 have been collected from MIG associated with the sequencing or from the closest validly published species.
Enzymes involved in the putative reductive acetyl-CoA pathway in T. ammonificans HB-1, closest relative homolog and homologs within the Aquificae phylum.
| Desulfurobac teriaceae | Hydrogenothe rmaceae | Aquificaceae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reaction | Putative enzyme | Putative gene locus | Closest relative | Putative origin‡ | D. thermolitho trophum | Desulfuro bacterium sp. TC5-1 | P. marina | Sulfurihydro genibium sp. YO3AOP1 | H. themophilus | Hydrogeno baculum sp. Y04AAS1H | Hydrogeni virga sp. 128–5 R1-6 | T. ruber | A. aeolicus |
| 1 | Formate dehydrogenase | Theam_1020 | Nitratiruptor sp. SB155-2 sim. 55% YP_001357016 | Methanogens | f.e.§ - 30% YP_00428203# | -** | 56% YP_002730364 | f.e.§ - 32% YP_001930236 | f.e.§ - 33% YP_003433330 | - | - | f.e.§ - 27% YP_003474076 | - |
| 2* | 5-formyltetra hydrofolate cyclo-ligase | Theam_1206 | Hydrogenivirga sp. 128–5 R1-1 sim. 41% WP_008285842 | Deltaproteo bacteria / Gram + | 41% YP_004281339 | 44% WP_022846876.1 | f.e.§ - 36% YP_002729868 | f.e.§ - 38% YP_001931834 | f.e.§ - 38% YP_003431710 | f.e.§ - 30% YP_002121539 | 41% WP_008285842 | 40% YP_003472981 | 43% 1SOU_A |
| formyltetra hydrofolate deformylase / hydrolase | Theam_0826 | Hydrogenivirga sp. 128–5 R1-1 sim. 70% WP_008287030 | Bacteria | 82% YP_004281077 | 77% WP_022846479 | - | - | - | - | 70% WP_008287030 | 65% NP_214247 | - | |
| phosphori bosylglycina mide formyl transferase 2 | Theam_1211 | P. marina sim. 76% YP_002731257 | Methanogens / Deltaproteo bacteria | 78% YP_004281335 | 64% WP_022847512 | 76% YP_002731257 | 73% YP_001931730 | f.e.§ - 28% YP_003433072 | 67% YP_007499479 | 74% WP_008288476 | f.e.§ - 25% YP_003473050 | f.e.§ - 30% NP_213168 | |
| phosphori bosylaminoi midazole carboxamide formyltransferase /IMP cyclohydrolase | Theam_0328 | P. marina sim. 59% YP_002731268 | Aquificae / Bacteria | 86% YP_004281681 | 78% WP_022847359 | 59% YP_002731268 | 56% YP_001931239 | 53% YP_003433270 | 50% YP_007499473 | 57% WP_008288360 | 52% YP_003474337 | 56% NP_214344 | |
| 3 | methylenetetra hydrofolate dehydrogenase / methenyltetra hydrofolate cyclohydrolase | Theam_1261 | A. aeolicus sim. 60% NP_214304 | Aquificae / Gram + | 87% YP_004281147 | 74% WP_022846217 | 55% YP_002731476 | 58% YP_001931240 | 59% YP_005511318 | 58% YP_001931240 | - | 58% YP_003473104 | 60% NP_214304 |
| 4 | methylenetetra hydrofolate reductase | Theam_0919 | Methanosrcina barkeri sim. 48% YP_305816 | Methanogens | 87% YP_004281147 | 74% WP_022846217 | 55% YP_002731476 | 58% YP_001931240 | 59% YP_005511318 | 58% YP_002121558 | f.e.§ - 24% WP_008286783 | 58% YP_003473104 | 60% NP_214304 |
| 5 | 5-methyltetra hydrofolate corrinoid/iron sulfurprotein methyltransferase | Theam_1184 | Clostridium glycolicum sim. 42% WP_018589788 | Gram + | 82% YP_004281484 | 72% WP_022846603 | f.e.§ - 31% YP_002729885 | f.e.§ - 38% YP_001930381 | f.e.§ - 37% YP_003432344 | f.e.§ - 34% YP_002122031 | f.e.§ - 38% WP_008287326 | f.e.§ - 36% YP_003473438 | f.e.§ - 34% NP_213375 |
| 6† | Carbon-monoxide dehydrogenase | Theam_1337 | Candidatus Methano peredens sp. BLZ1 sim. 56% KPQ43483 | Archaea / Gram + | 87% WP_013638427 38% WP_013638030 | 76% WP_022847143 | 41% YP_002729904 | - | - | - | - | - | - |
-
* – Reactions are numbered according to Figure 1. For reaction two we reported the possible enzymes that could substitute the missing 10-fomyl-THF synthetase (Fhs). The enzymes are listed in order of decreasing likelihood of their involvement in the reaction based on putative substrate affinity.
-
† – Reaction six is catalyzed by the Acs/CodH complex, reported here separately.
-
‡ – Putative origin of the gene was calculated as consensus taxonomic assignment of the first 100 bastp hits against the nr database. Double assignment implies equal number of assignments to the two taxonomic groups.
-
§ – f.e. = functional equivalent annotated in the genome with similarity below 40% with T. ammonificans equivalent gene. Not considered a true homolog in the present study.
-
# – Pairwise similarity to T. ammonificans translated gene and accession number of the homologs.
-
** – Missing homolog or functional annotated equivalent in the genome.
List of the genomes belonging to the Aquificae phylum used for comparative genomic analyses.
| Organism | Genome acc. number | Genome length | GC content | Num. genes |
|---|---|---|---|---|
| Aquifex aeolicus VF5 | NC_000918 | 1,590,791 | 43% | 1782 |
| Desulfurobacterium sp. TC5-1 | NZ_ATXC01000001 | 1,653,625 | 40% | 1680 |
| Desulfurobacterium thermolithotrophum DSM 11699 | NC_015185 | 1,541,968 | 35% | 1561 |
| Hydrogenivirga sp. 128–5 R1-1 | NZ_ABHJ01000551 | 3,038,240 | 44% | 3756 |
| Hydrogenobacter thermophilus TK-6 | NC_013799 | 1,743,135 | 44% | 1897 |
| Hydrogenobaculum sp. Y04AAS1 | NC_011126 | 1,559,514 | 35% | 1631 |
| Persephonella marina EX-H1 | NC_012440 | 1,983,966 | 37% | 2067 |
| Persephonella sp. IF05-L8 | NZ_JNLJ01000001 | 1,828,858 | 35% | 1920 |
| Sulfurihydrogenibium azorense Az-Fu1 | NC_012438 | 1,640,877 | 33% | 1722 |
| Sulfurihydrogenibium sp. YO3AOP1 | NC_010730 | 1,838,442 | 32% | 1832 |
| Sulfurihydrogenibium subterraneum DSM 15120 | NZ_JHUV01000001 | 1,610,181 | 32% | 1701 |
| Sulfurihydrogenibium yellowstonense SS-5 | NZ_ABZS01000228 | 1,534,471 | 33% | 1637 |
| Thermocrinis albus DSM 14484 | NC_013894 | 1,500,577 | 47% | 1631 |
| Thermocrinis ruber DSM 23557 | NZ_CP007028 | 1,521,037 | 45% | 1625 |
| Thermocrinis sp. GBS | NZ_JNIE01000001 | 1,315,625 | 41% | 1417 |
| Thermosulfidibacter takaii ABI70S6 | NZ_AP013035 | 1,816,670 | 43% | 1844 |
| Thermovibrio ammonificans HB-1 | NC_014926 | 1,759,526 | 52% | 1820 |





