Suppression of C9orf72 RNA repeat-induced neurotoxicity by the ALS-associated RNA-binding protein Zfp106
Figures
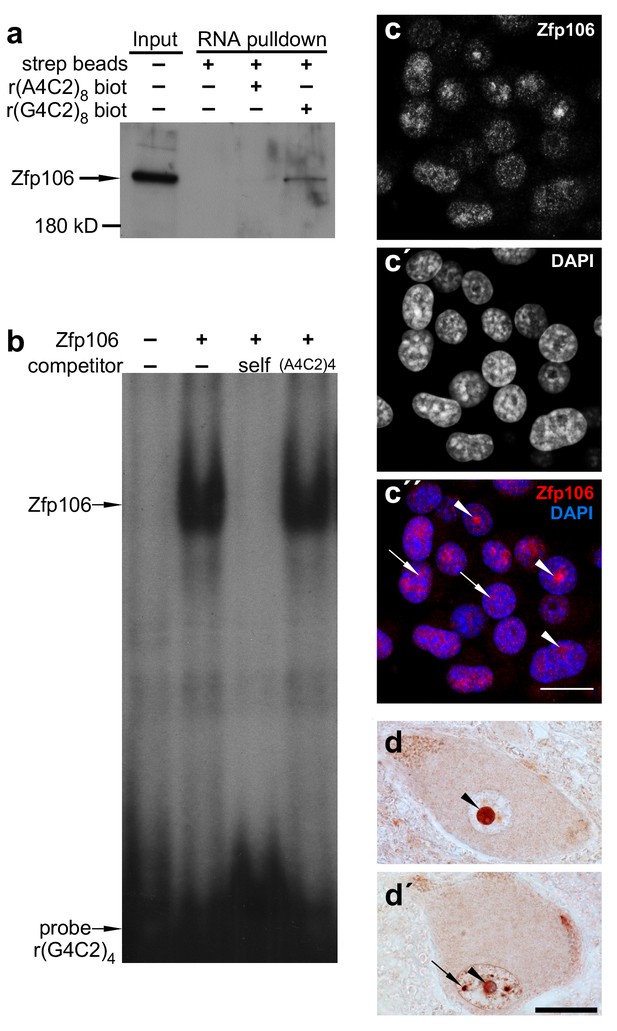
Zfp106 is an rGGGGCC binding protein.
(a) Western blot analysis of Zfp106 eluted from a biotinylated-RNA pulldown of Neuro-2a nuclear proteins. Endogenous Zfp106 was specifically pulled down by (GGGGCC)8 biotinylated RNA and not by the unrelated (AAAACC)8 biotinylated RNA oligonucleotide. (b) RNA EMSA performed with purified Zfp106 protein demonstrates that Zfp106 directly binds (GGGGCC)4 RNA in vitro. 30× molar excess of unlabeled self competitor, but not 30× r(AAAACC)4, competes with Zfp106 binding to r(GGGGCC)4, establishing specificity of the interaction. (c) Immunofluorescence with an anti-Zfp106 antibody detects Zfp106 expression in the nucleolus (arrowheads) and in other discrete nuclear puncta (arrows) in cultured Neuro-2a cells. The three images are the same section showing the red channel in (c), the blue channel in (c′), and both channels in (c′′). Scale bar, 20 μm. (d, d′) Immunohistochemical detection of human ZNF106 shows strong nucleolar expression in human motor neurons in the anterior horn of spinal cord. The two images are from different post-mortem individuals. Arrowheads mark nucleolar ZNF106 expression; arrows mark other nuclear and perinucleolar foci of ZNF106 expression. Scale bar, 25 µm.
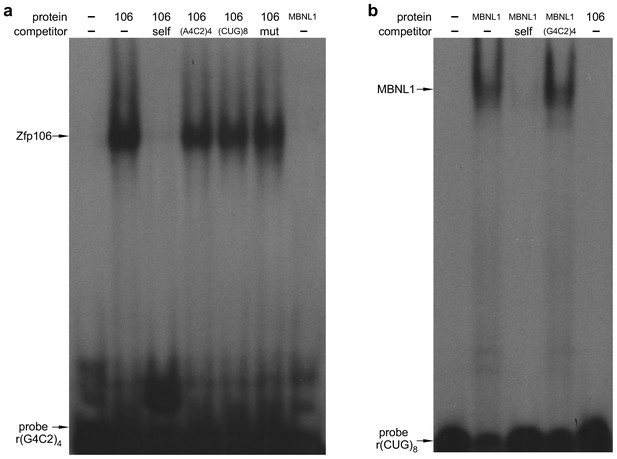
Zfp106 binds specifically to rGGGGCC repeats.
(a) RNA EMSA performed with purified Zfp106 protein demonstrates that Zfp106 binds specifically to r(GGGGCC)4in vitro since binding is efficiently competed by 30× molar excess of unlabeled self competitor but not 30× molar excess of r(AAAACC)4, r(CUG)8 or a mutated r(GGGGCC)4 sequence [mut, r(GAGGCCGGGACCGAGACCGAGGCC)]. Purified MBNL1 protein was used as a negative control for r(GGGGCC)4 binding. MBNL1 does not bind to r(GGGGCC)4 under the same conditions in which Zfp106 shows efficient binding. (b) RNA EMSA using purified Zfp106 protein demonstrates that Zfp106 does not bind r(CUG)8 repeats under conditions in which the RNA binding protein MBNL1 efficiently binds r(CUG)8. MBNL1 binding to r(CUG)8 is specific since binding is competed by 30× molar excess of self competitor but not 30× molar excess of r(GGGGCC)4.
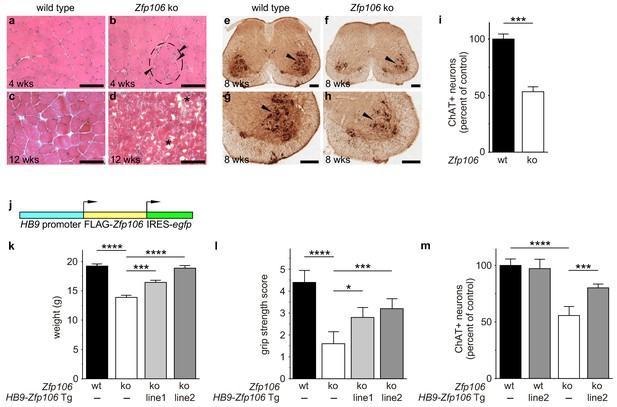
Zfp106 loss-of-function causes a severe neuromuscular phenotype in mice.
(a–h) Evidence of profound neuromuscular pathology in Zfp106-null mice. H&E stained sections of quadriceps muscles (a–d) from 4-week old and 12-week old wild type (a, c) and Zfp106 knockout (b, d) mice are shown. Note the presence of grouped small angular fibers in knockout mice (dashed circle), already clearly evident by 4-weeks of age (b, arrowheads) compared to the rounder, more evenly-shaped fibers in controls (a). By 12-weeks (end stage disease), Zfp106 knockout mice have profound pathology and the presence of atrophic and degenerated muscle fibers with numerous inclusions (d, asterisks). Scale bar, 100 μm. ChAT immunohistochemical staining (e–h) of lumbar spinal cord sections from 8-week old wild type (e, g) and Zfp106 knockout (f, h) mice show a drastic reduction in the number of ChAT+ motor neurons. Arrowheads mark ChAT-expressing motor neurons in the ventral horn. Scale bar, 200 μm in all panels. (i) Quantification of ChAT-positive neurons in the lumbar spinal cord of wild type and Zfp106 knockout mice at 8 weeks of age shows a ~ 47% loss of motor neurons in Zfp106 knockout mice. n = 5 mice for each genotype. Data are expressed as percent of control ± SD and were analyzed by t-test. ***p<0.001. (j) Schematic of the HB9-3×FLAG-2×STREP-Zfp106-IRES-gfp (HB9-Zfp106) transgene. (k, l, m) Transgenic expression of Zfp106 in motor neurons suppresses the wasting phenotype (k), the reduction in grip strength (l) and the loss of lumbar spinal cord motor neurons (m) in Zfp106 knockout mice. Wasting data are expressed as the mean weight ± SEM. Grip strength data are expressed as the mean grip score ± SD; grip score: 1, 1–10 s; 2, 11–25 s; 3, 26–60 s; 4, 61–90 s; 5, > 90 s. ChAT-positive neuron counts are expressed as percent of control ± SD. n = 5 female mice at 8 weeks of age for each genotype. Data were analyzed by one-way ANOVA and Bonferroni's Multiple Comparison Test. *p<0.05; ***p<0.001, ****p<0.0001.
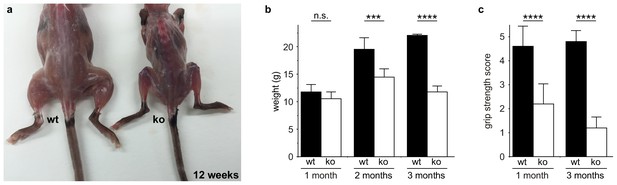
Zfp106 knockout mice exhibit progressive and severe weight and grip strength loss over time.
(a) Dorsal view of skinned wild type and Zfp106 knockout mice at 12-weeks of age. Zfp106 knockout mice show severe muscle wasting and kyphosis compared to wild type mice. (b) Female mice were weighed at the indicated ages, n = 5 for each group. Body weight of Zfp106 knockout mice was significantly reduced at 2 months of age compared to control and further reduced at 3 months of age. (c) Zfp106-null mice exhibit profound loss of grip strength; grip score: 1, 1–10 s; 2, 11–25 s; 3, 26–60 s; 4, 61–90 s; 5, > 90 s. n = 5 age-matched female mice for each group tested at the indicated ages. Graphical data are presented as the mean ± SD. Data were analyzed by one-way ANOVA and Bonferroni's Multiple Comparison Test. ns, not significant; ***p<0.001, ****p<0.0001.
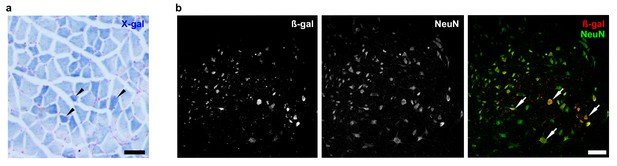
Zfp106 is expressed in skeletal muscle and motor neurons.
Expression of Zfp106 as detected by β-galactosidase activity in quadriceps (a) and spinal cord (b) of 8-week-old Zfp106laczflox/+ mice. β-galactosidase is expressed from a lacZ knock-in at the Zfp106 locus. A section of the quadriceps muscle from an 8-week old Zfp106laczflox/+ mouse stained with X-gal is shown in (a). Arrowheads show β-galactosidase-expressing muscle fibers, weakly detectable by X-gal staining; scale bar, 100 μm. A section through the ventral horn of the spinal cord (lumbar region) of a Zfp106laczflox/+ mouse is shown in (b). The same section is shown in all three panels in (b); the green channel shows expression of the neuronal marker NeuN; the red channel shows expression of β-galactosidase expressed from the Zfp106 locus. Arrows denote co-expression of β-galactosidase and NeuN in large motor neurons in the ventral horn of the spinal cord; scale bar, 50 μm.
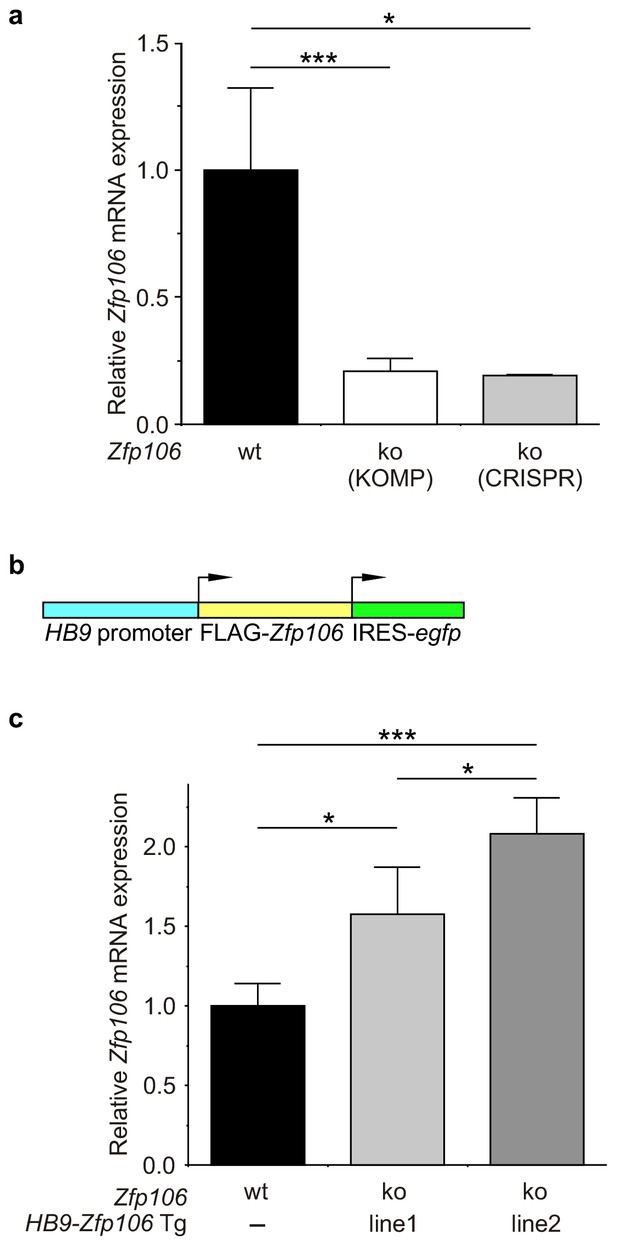
Expression of Zfp106 in wild type, knockout and HB9-Zfp106 transgenic mice.
(a) Evaluation of Zfp106 expression by RT-qPCR in lumbar spinal cord of wild type, KOMP Zfp106 knockout and CRISPR Zfp106 knockout mice. Expression of Zfp106 mRNA is drastically and significantly reduced in lumbar spinal cord of both KOMP and CRISPR knockout mice. (b) Schematic of the transgenic construct used for generation of the motor neuron specific Zfp106 mice. The HB9 promoter drives expression of a 3xFLAG-2xSTREP N-terminally tagged Zfp106 and IRES-EGFP. (c) Evaluation of transgene expression by RT-qPCR in lumbar spinal cord at P10. Expression of Zfp106 mRNA ranged from ~1.5 to 2 fold of wild type expression in two independent transgenic mouse lines, as assessed by qPCR with primers amplifying both endogenous and transgenic Zfp106. Data in a, c were analyzed by one-way ANOVA and Bonferroni's Multiple Comparison Test. *p<0.05; ***p<0.001.
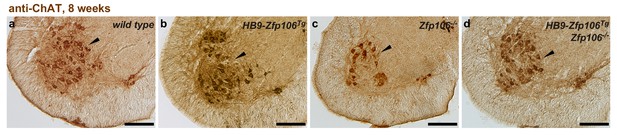
Rescue of ChAT+ motor neuron loss by restoration of Zfp106 to motor neurons in Zfp106 knockout mice.
Representative images of transverse sections of lumbar spinal cord from wild type (a), motor neuron-specific Zfp106 transgenic (b), Zfp106 knockout (c), and Zfp106 knockout/motor neuron-specific Zfp106 transgenic mice (d), stained by anti-ChAT antibody. Arrowheads mark ChAT+ motor neurons. Scale bar, 200 µm.
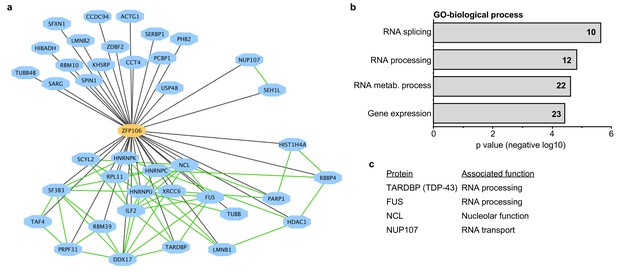
Zfp106 interacts with a network of other ALS-associated RNA-binding proteins.
(a) Network representation of the Zfp106 interactome generated by Cytoscape. Zfp106 (yellow node) interacts with 39 proteins with top 5% MiST score (blue nodes). Curated protein-protein interactions from the CORUM database are indicated as nodes connected by green lines. (b) Gene Ontology (GO) Biological Process analysis of the Zfp106 interactome ranked by p value. The number of interacting proteins (top 5% by MiST score) in each category is indicated inside each bar. (c) Selected ALS-associated proteins within the top 5% MiST-scoring hits from the Zfp106 interactome are listed with putative functions associated with the pathobiology of ALS indicated.
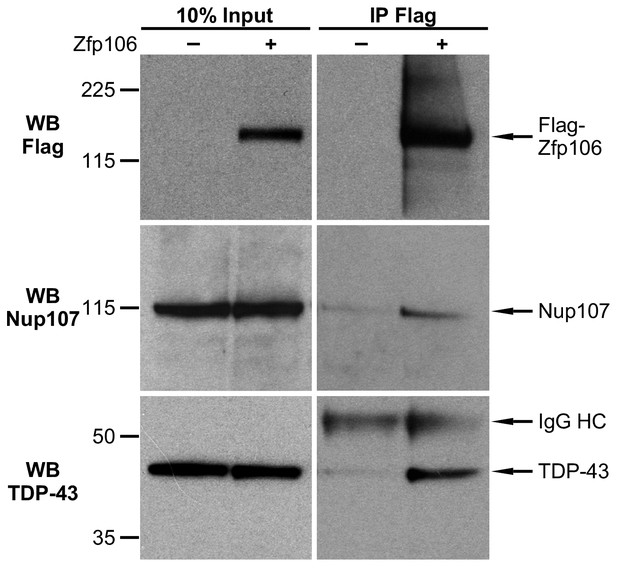
Zfp106 interacts with TDP43 and Nup107 in Neuro-2a cells.
Endogenous TDP43 and Nup107 coimmunoprecipitated with Zfp106 in Neuro-2a cells. Cells were transfected with 3×FLAG-2×STREP-Zfp106 or 3×FLAG-2×STREP parental vector and lysates were subjected to immunoprecipitation with anti-FLAG beads, followed by anti-FLAG (top), anti-Nup107 (middle), and anti-TDP43 (bottom) western blotting. Nup107 and TDP43 were coimmunoprecipitated by anti-FLAG IP only in lysates from cells expressing FLAG-Zfp106. The immunoglobulin heavy chain (IgG HC) of the anti-FLAG antibody is indicated. Molecular weight markers in kD are indicated on the left.
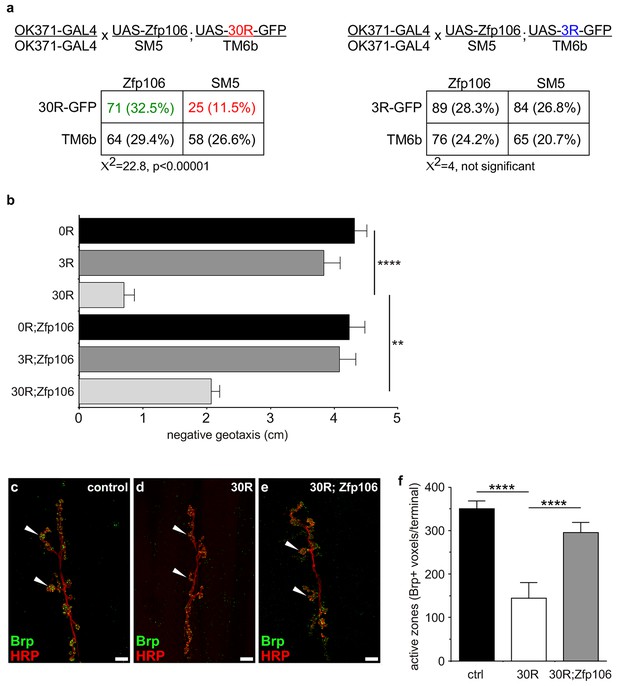
Zfp106 suppresses GGGGCC-mediated neurotoxicity in Drosophila.
(a) Schematics and Punnett squares from crosses of UAS-Zfp106; UAS-30R-GFP (left) and UAS-Zfp106; UAS-3R-GFP (right) transgenic flies to OK371-GAL4 flies. The Punnett squares show the number of flies that eclosed from the indicated crosses and show that expression of 30R-GFP resulted in failure of ~50% of flies to eclose (25 actual versus 54.5 predicted); this defect was completely suppressed by co-expression of Zfp106. The Chi-square test (χ2) was used to determine the significance of the differences between the observed and expected frequencies of the expected genotypes. (b) Locomotor activity of 28-day post-eclosion flies of the indicated genotypes assessed by rapid iterative negative geotaxis (RING) assay. Data are presented as the mean height climbed ± SEM. A two-way ANOVA analysis of the locomotor data revealed a significant effect of repeat expression (p<0.0001) and Zfp106 expression (p<0.01) on locomotor activity, with a significant interaction between the two factors (p<0.01); i.e. Zfp106 coexpression significantly suppresses neurotoxicity induced by expression of 30R (p<0.01) by Bonferroni's posthoc Multiple Comparison Test. n = 25 male flies for each genotype; ****p<0.0001, **p<0.01. (c, d, e) Representative NMJs of muscle 6/7 in abdominal segment A3 of third instar larvae of the indicated genotypes co-stained with anti-HRP (neuronal marker, red channel) and anti-Bruchpilot (Brp) (active zone marker, green channel). Staining of the active zone component Brp shows that expression of 30R causes a reduction in Brp-positive active zones [arrowheads in (d) compared to control in c]. Coexpression of Zfp106 suppressed the reduction in Brp-positive active zones caused by 30R [arrowheads in e compared to d]. (f) Quantification of active zones as total voxel occupancy by Brp signal at each terminal shows that Zfp106 significantly suppresses the reduction in active zone caused by expression of 30R. Data were analyzed by one-way ANOVA and Bonferroni's Multiple Comparison Test. ***p<0.001, ****p<0.0001.
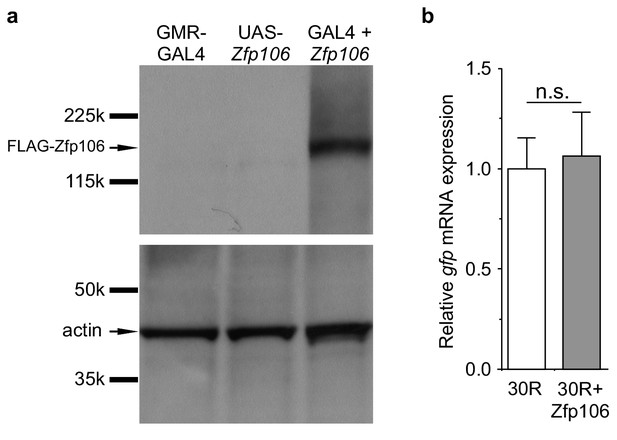
GAL4-dependent expression of Zfp106 in Drosophila does not affect 30R-gfp mRNA levels.
(a) Anti-FLAG western blot conducted on lysates from heads dissected from transgenic flies of the indicated genotypes. The band in lane 3 migrating below the 225 kD marker indicates that UAS-3×FLAG-2×STREP-Zfp106 (UAS-Zfp106) transgenic flies express Zfp106 in a GAL4-dependent manner. (b) Evaluation of 30R-gfp expression by RT-qPCR in flies expressing 30R alone or coexpressing 30R and Zfp106. Zfp106 coexpression does not cause alterations in 30R-gfp mRNA levels.
Videos
Zfp106-null mice exhibit a severe progressive neurodegenerative phenotype.
The video shows a Zfp106 knockout mouse and a littermate control at 10 weeks of age. The Zfp106 knockout displays abnormal gait due to hind limb paralysis, severe kyphosis, wasting, and persistent hind limb clasping at tail suspension. The wild type mouse displays normal gait and normal extension of the hind limbs away from the abdomen during the tail suspension test.
Additional files
-
Supplementary file 1
Zfp106 interactors identified in HEK293T cells.
39 proteins (indicated by Uniprot accession and gene name) identified with 95% confidence based on MiST score by affinity-purification mass spectrometry of FLAG-Zfp106 in HEK293T cells.
- https://doi.org/10.7554/eLife.19032.015
-
Supplementary file 2
Protein complexes overrepresented in the Zfp106 interactome.
List of protein-protein pairs in protein complexes from the CORUM database that are overrepresented in the Zfp106 interactome. Proteins are indicated by Uniprot accession number and gene name, protein complexes by CORUM protein complex ID and name.
- https://doi.org/10.7554/eLife.19032.016





