Notch signaling and Bsh homeodomain activity are integrated to diversify Drosophila lamina neuron types
Figures

Notch signaling is activated in newborn L4 neurons but not L5.
(A) Schematic of Notch signaling pathway. (B-B”) Hey as a reporter of active Notch signaling is only expressed in newborn L4 neurons but no other lamina neurons at 15 hr APF. Here and below, scale bar: 10 µm, n≥5 brains. Dashed line delineates the boundary between Bsh+ L4 and Bsh+ L5 neurons. (C, D) Hey is expressed prior to the activation of the secondary HDTFs Ap and Pdm3 at 15 hr APF. The dashed line delineates the boundary between Bsh+ L4 and Bsh+ L5 neurons. (E–H) Using twin-spot MARCM, two sibling neurons generated by one progenitor are traced. RFP and GFP cells are either both L4 neurons (Bsh+Pdm3-) or both L5 neurons (Bsh+Pdm3+). N=4. The dashed line outlines RFP+ and GFP+ cell bodies.
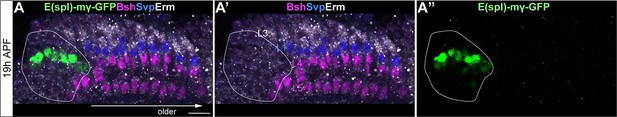
E(spl)-mγ is not expressed in lamina neurons.
(A-A”) E(spl)-mγ-GFP is expressed in the LPC region but not in lamina neurons. Erm labels L3; Svp labels L1; Bsh labels Bsh+ LPCs, L4 and L5. A solid white line outlines the LPC region.
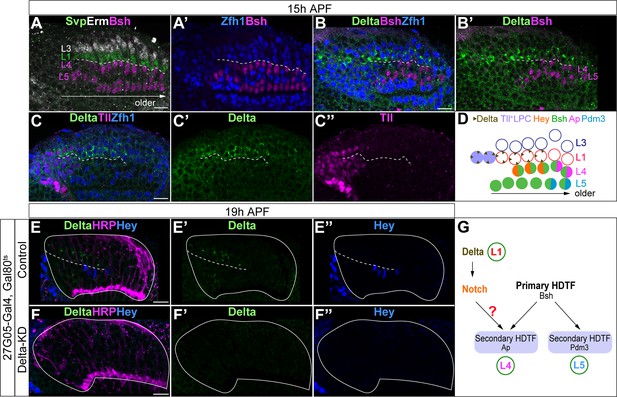
L1 neurons express Delta and activate Notch signaling in adjacent L4 neurons.
(A, A’) Svp+Zfh1+L1 neurons are adjacent to both newborn L3 and L4 neurons. Newborn L3 neurons (Erm+) are localized strictly above (distal to) L1 neurons (Svp+Zfh1+), and L1 neurons are localized strictly above (distal to) L4 neurons. Here and below, scale bar: 10 µm, n≥5 brains. The dashed line delineates the boundary between L1 (Svp+Zfh1+) and L4 (Bsh+) cell bodies. (B, B’) Delta is expressed in Zfh1+ L1 neurons which are adjacent to Bsh+ L4 neurons. The dashed line delineates the boundary between L1 (Zfh1+) and L4 (Bsh+) cell bodies. (C-C”) Delta is also expressed in a subset of LPCs (Tll+). The dashed line highlights Delta+ cell bodies. (D) Summary of A-C data; triangles represent Delta expression. (E-F”) Delta-KD (27G05-Gal4, tubP-GAL80[ts], UAS-Delta-RNAi) results in loss of Delta and Hey expression in lamina. HRP labels the axons of the photoreceptors, which represent the lamina column. A solid white line outlines the lamina and a dashed line delineates the boundary between Delta+ cells and Hey+ cells. (G) Summary.
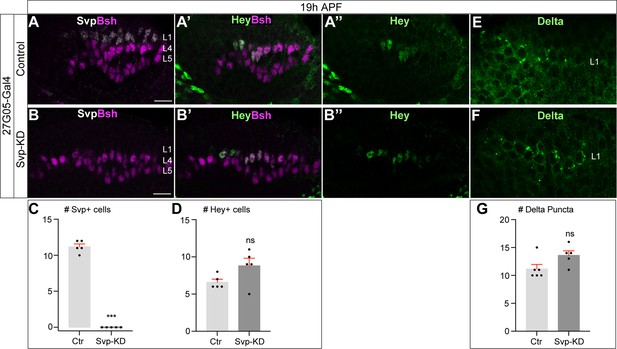
Svp is not required to initiate or maintain Delta expression in L1 neurons.
(A–D) In Svp-KD (27G05-Gal4>UAS-Svp-RNAi), there is a loss of Svp while Hey expression remains unaffected at 19 hr APF. (C–D) Quantification (single optical section). Here and below, scale bar, 10 µm, n≥5 brains. (E–G) In Svp-KD (27G05-Gal4>UAS-Svp-RNAi), Delta expression remains unaffected at 19 hr APF. (G) Quantification (single optical section). Data are presented as mean ± SEM. Each dot represents each brain. n=5 brains in (C) and (D). n=6 brains for control and n=5 brains for Svp-KD in (G). ***p<0.001, ns = not significant, unpaired t-test.
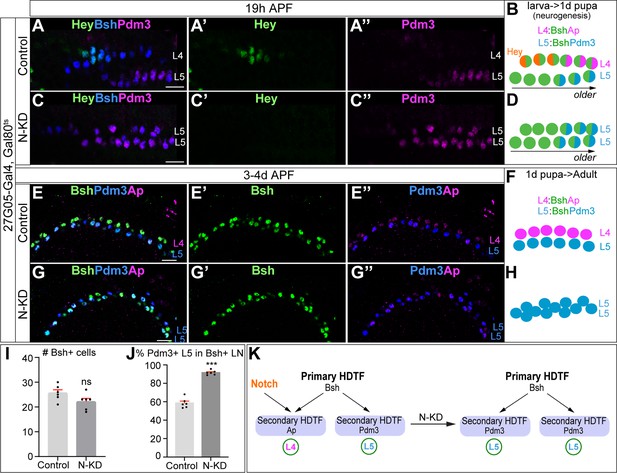
Bsh without Notch signaling activates Pdm3 and specifies L5 neuronal fate.
(A–D) N-KD in lamina (27G05-Gal4, tubP-GAL80[ts], UAS-Notch-RNAi) shows Hey expression is absent, and Bsh only activates Pdm3 and specifies L5 neuronal fate during lamina neurogenesis (19 hr APF). Here and below, scale bar, 10 µm, n≥5 brains. (E–J) N-KD in lamina shows Bsh+ lamina neurons are mainly Pdm3+ L5 neurons, though the number of Bsh+ lamina neurons remains unaffected at 3-4d APF. (I–J) Quantification (single optical section). (K) Summary Data are presented as mean ± SEM. Each dot represents each brain. n=6 brains in (I), (J). ***p<0.001, ns = not significant, unpaired t-test.
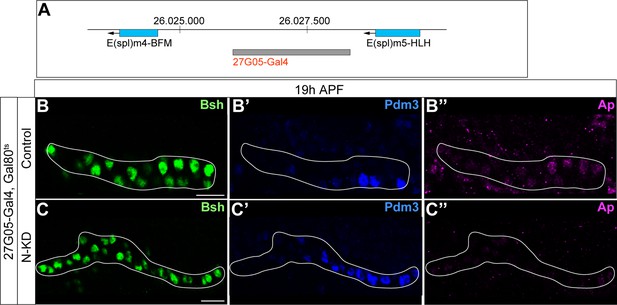
27G05-Gal4 is inserted in between two Notch target genes; Bsh without Notch signaling activates Pdm3 and specifies L5 morphology.
(A) Schematic of 27G05-Gal4 insertion site in between two Notch target genes (E(spl)m4-BFM and E(spl)m5-HLH). (B-C”) Bsh without Notch signaling activates Pdm3 and specifies L5 morphology. In N-KD (27G05-Gal4, tubP-GAL80[ts], UAS-Notch-RNAi), Bsh activates Pdm3 but not Ap and specifies L5 neuronal fate during lamina neurogenesis (19 hr APF). The white line outlines Bsh+ lamina cells. Scale bar, 10 µm, n≥5 brains.
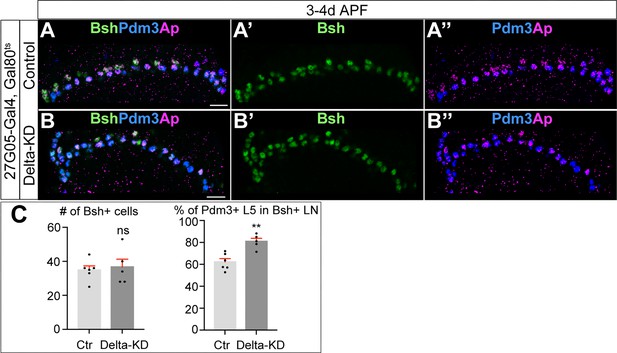
Bsh specifies L5 neuronal fate over L4 following Delta-KD.
(A–C) Delta-KD (27G05-Gal4, tubP-GAL80[ts], UAS-Delta-RNAi) in lamina shows the increase of Pdm3+ L5 neurons at the expense of L4 at 3-4d APF. (C) Quantification (single optical section). Scale bar, 10 µm. Data are presented as mean ± SEM. Each dot represents each brain. n=5 brains. **p<0.01, ns = not significant, unpaired t-test.
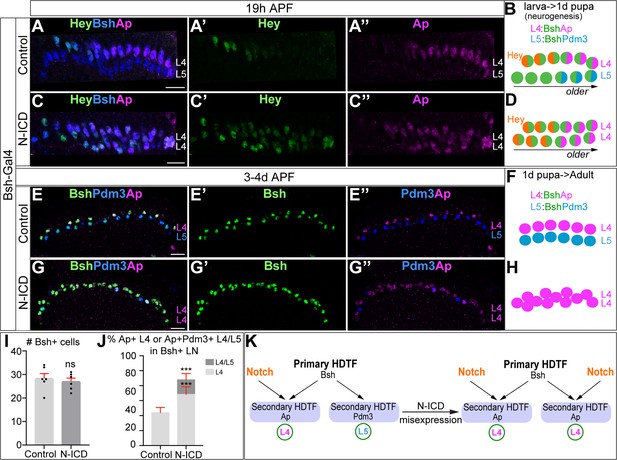
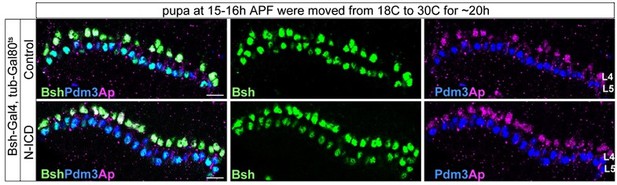
Bsh with Notch signaling activates Ap and specifies L4 neuronal fate.
(A–D) Ectopic expression of N-ICD in newborn L5 neurons (Bsh-Gal4 >UAS-N-ICD) results in ectopic Hey and Ap activation and an increased number of L4 neurons at 19 hr APF. Here and below, scale bar, 10 µm, n≥5 brains. (E–J) N-ICD shows Bsh+ lamina neurons are mainly Ap+ L4 neurons, though the number of Bsh+ lamina neurons remains unaffected at 3-4d APF. (I–J) Quantification (single optical section). (K) Summary. Data are presented as mean ± SEM. Each dot represents each brain. n=6 brains in (I), (J). ***p<0.001, ns = not significant, unpaired t-test.
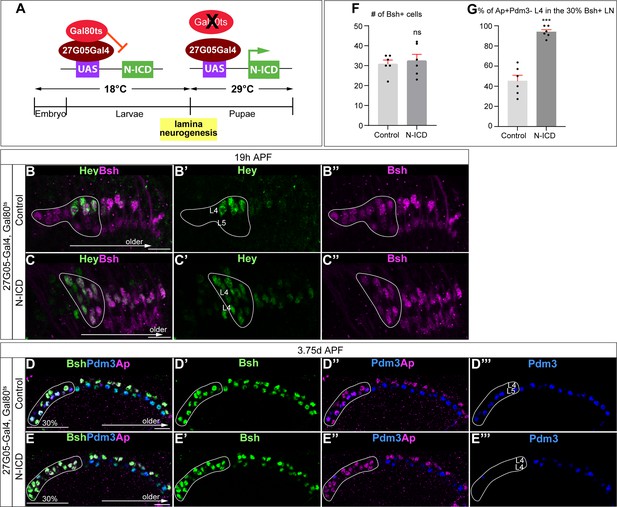
Temporally restricted activation of Notch signaling by 27G05-Gal4 enables Bsh to specify L4 neuronal fate over L5.
(A) Schematic of the genetics for temporally restricted activation of Notch signaling. (B-C”) Activation of Notch signaling from 0 hr APF (affected region circled) results in ectopic Hey expression at 19 hr APF. Here and below, scale bar, 10 µm, n≥5 brains. (D–G) Activation of Notch signaling from 0 hr APF (affected region circled) results in no change to Bsh expression, loss of Pdm3+ L5 neurons, and gain of Ap+Pdm3 L4 neurons at 3-4d APF. (F and G) Quantification (single optical section). Data are presented as mean ± SEM. Each dot represents each brain. ***p<0.001, ns = not significant, unpaired t-test.
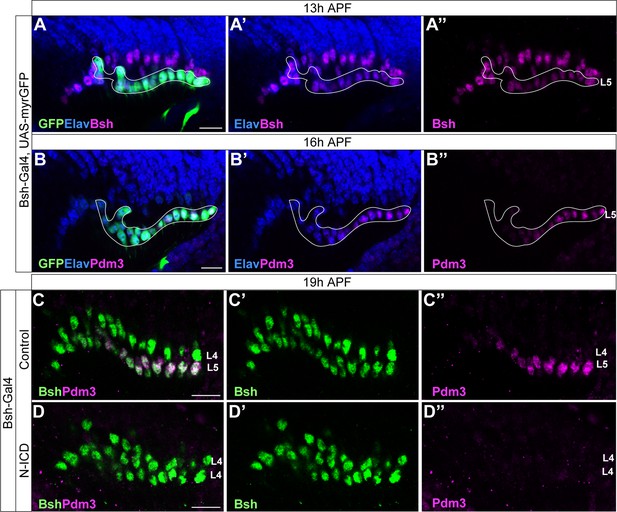
Bsh-Gal4 is expressed in newborn L5 neurons; Bsh does not activate L5 marker Pdm3 when Notch signaling is activated in newborn L5 by Bsh-Gal4 and UAS-N-ICD.
(A-B”) Bsh-Gal4 is expressed in newborn L5 neurons. (A-A”) GFP+ cells (white line circle; Bsh-Gal4 >UAS-myrGFP) label L5 neurons which are in the bottom (proximal) row of Bsh+ cells during lamina neurogenesis (13 hr APF). Here and below, scale bar, 10 µm, n≥5 brains. (B-B”) GFP+ cells (white line circle; Bsh-Gal4 >UAS-myrGFP) label Pdm3+ newborn L5 and early-born L5 neurons during lamina neurogenesis (16 hr APF). (C-D”) Bsh does not activate L5 marker Pdm3 when Notch signaling is activated in newborn L5 by Bsh-Gal4 and UAS-N-ICD. Pdm3 expression is initiated in L5 neurons in control but not in N-ICD (Bsh-Gal4 >UAS-N-ICD).
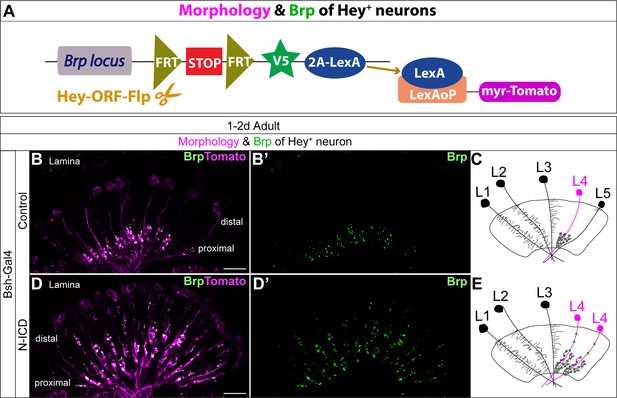
Bsh-Gal4 activation of Notch signaling in newborn L5 neurons specifies L4-like morphology and presynaptic sites.
(A) Schematic of the genetics used to trace the morphology and presynaptic site (Brp) of Hey+ neurons. (B–E) In controls, Tomato+ neurons in lamina have L4 morphology and presynaptic sites labeled by Brp-V5. In Bsh-KD, Tomato+ cells, which trace the endogenous and ectopic Hey+ neurons, have L4-like morphology and presynaptic sites labeled by Brp-V5 in the lamina. Scale bar, 10 µm. n≥5 brains.
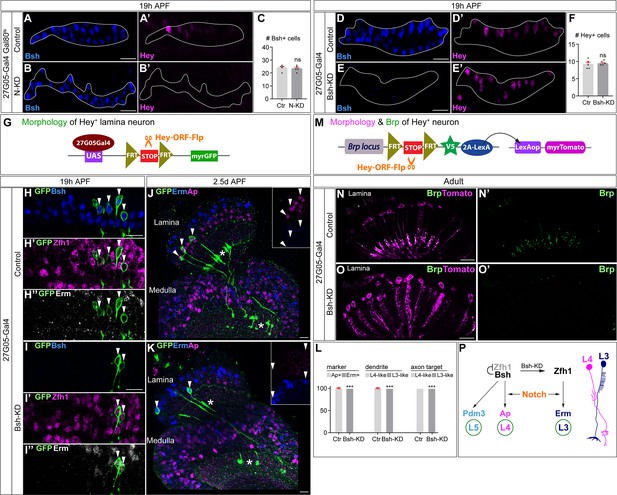
Notch activation and Bsh expression are mutually independent; Notch signaling without Bsh specifies L3 neuron type.
(A–F) Notch activation and Bsh expression are mutually independent. (A–C) N-KD in lamina (27G05-Gal4, tubP-GAL80[ts], UAS-Notch-RNAi) results in loss of Hey expression without affecting Bsh expression. (C) Quantification (single optical section). Here and below, scale bar, 10 µm, n≥5 brains. (D–F) Bsh-KD in lamina (27G05-Gal4>UAS-Bsh-RNAi) results in loss of Bsh expression without affecting Hey expression. (F) Quantification (single optical section). (G–L) Notch signaling without Bsh specifies L3 neuron type. (G) Schematic of the genetics used to trace the morphology of Hey+ lamina neurons. (H, I) In controls, GFP+ neurons express the L4 marker Bsh (white arrowhead) at 19 hr APF. In Bsh-KD, Bsh expression becomes absent in lamina and GFP+ cells now express L3 markers Zfh1 and Erm (white arrowhead). (J–L) In controls, GFP + cells express the L4 marker Ap (white arrowhead) and have L4 morphology (asterisk) at 2.5d APF. In Bsh-KD, GFP+ cells express L3 marker Erm (white arrowhead) and adopt L3 morphology (asterisk). (L) Quantification for (J) and (K) (single optical section). (M–O) (M) Schematic of the genetics used to trace the morphology and presynaptic sites of Hey+ neurons. (N, O) In controls, Tomato+ neurons have L4 morphology and presynaptic sites (Brp) in the lamina. In Bsh-KD, Tomato+ neurons adopt L3 morphology and connectivity, which lacks presynaptic localization in the lamina (Xu et al., 2019). (P) Summary. Data are presented as mean ± SEM. Each dot represents each brain. n=5 brains in (C), (F), and (L). ***p<0.001, ns = not significant, unpaired t-test.
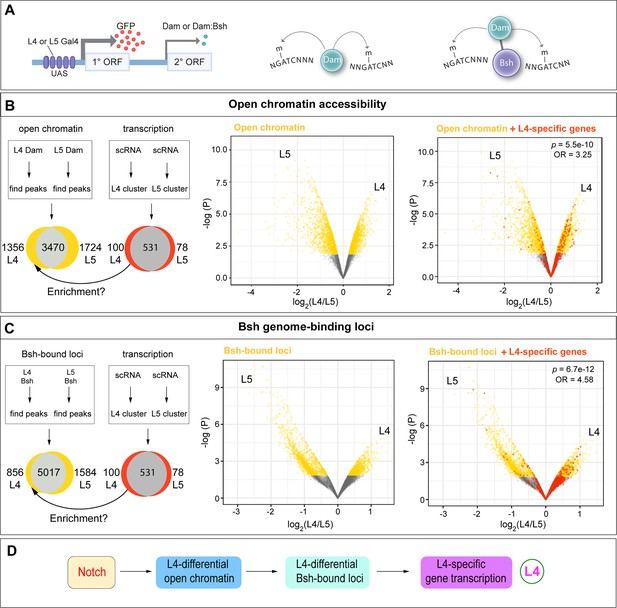
The NotchON L4 has correlated open chromatin, Bsh-bound loci, and transcription profile that is distinct from the NotchOFF L5.
(A) Schematic of the Targeted DamID method. Upon GAL4 induction, low levels of either Dam or Bsh:Dam are expressed, allowing genome-wide open chromatin and Bsh binding targets to be identified. (B) L4 and L5 neurons show distinct open chromatin regions (yellow), and L4-specific transcribed genes (red) are enriched in L4-distinct open chromatin regions. (C) L4 and L5 neurons show distinct Bsh-bound loci (yellow), and L4-specific transcribed genes (red) are enriched in L4-distinct Bsh-bound loci. p Values from Fisher’s exact test; odds ratios (OR) expressed as L4/L5 in all cases. (D) Model.
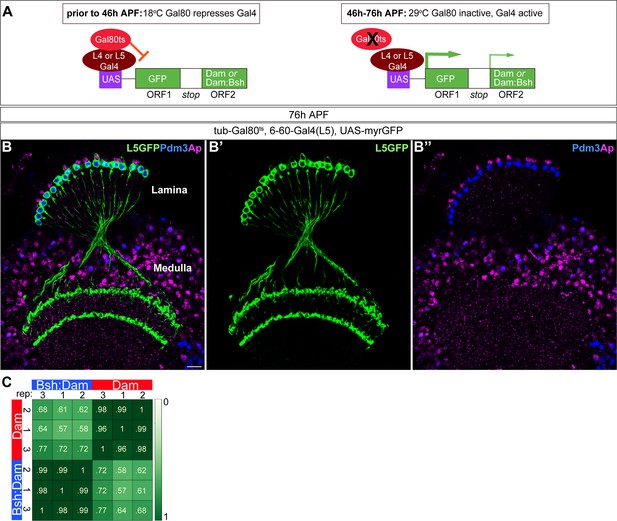
Six to 60 Gal4 used for TaDa experiment is specifically expressed in L5 neurons at 46 –76 hr APF.
(A) Schematic of the genetics used to perform targeted Dam ID experiments (see Figure 6). (B-B”) 6–60 Gal4, which is expressed at 46–76 hr APF, drives GFP expression in all Pdm3+ L5 neurons and weak GFP expression in some Ap+ L4 neurons. Scale bar, 10 µm. n≥5 brains. (C) Both Dam and Bsh:Dam show high Pearson correlation coefficients among their three biological replicates, with lower Pearson correlation coefficients between Dam and Bsh:Dam. Heat map is generated using the multiBamSummary and plotCorrelation functions of deepTools.
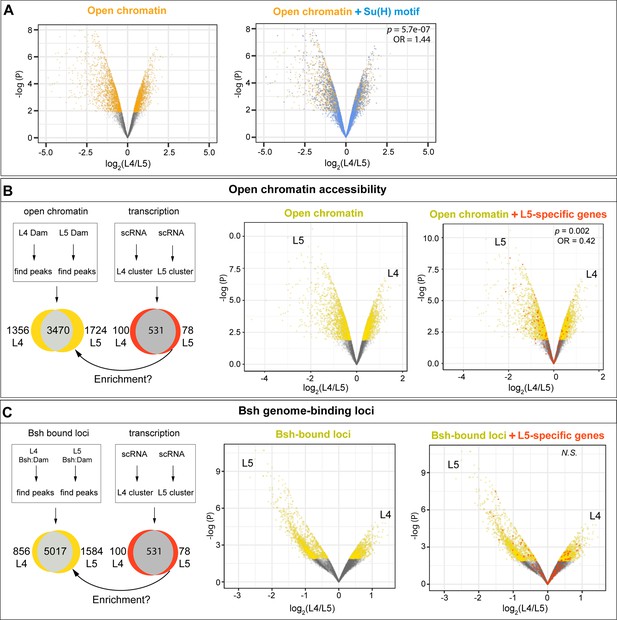
L4-distinct open chromatin regions are more likely to contain the Su(H) binding motif; L5-specific transcribed genes are correlated with L5-distinct open chromatin regions but not L5-distinct Bsh-bound loci.
(A) L4 and L5 neurons show distinct open chromatin regions (yellow), and the Su(H) binding motif (blue) is more likely to be present in L4-distinct open chromatin regions. (B) L4 and L5 neurons show distinct open chromatin regions (yellow), and L5-specific transcribed genes (red) are enriched in L5-distinct open chromatin regions. (C) L4 and L5 neurons show distinct Bsh-bound loci (yellow); L5-specific transcribed genes (red) and L5-distinct Bsh-bound loci do not show correlation. p Values from Fisher’s exact test; odds ratios (OR) expressed as L4/L5 in all cases.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (D. melanogaster) | 10xUAS-IVS- myristoylated-GFP | Bloomington Drosophila Stock Center | RRID: BDSC_32199 | w[1118]; P{y[+t7.7] w[+mC]=10XUAS-IVS-myr::GFP}su(Hw)attP5 |
| Strain, strain background (D. melanogaster) | R27G05GAL4 | Bloomington Drosophila Stock Center | RRID: BDSC_48073 | w[1118]; P{y[+t7.7] w[+mC]=GMR27 G05-GAL4}attP2 |
| Strain, strain background (D. melanogaster) | UAS-Bsh-RNAi | Bloomington Drosophila Stock Center | RRID: BDSC_29336 | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02498}attP2 |
| Strain, strain background (D. melanogaster) | yw; UAS-mCD8-GFP, UAS-rCD2i, FRT40A/ CyO; TM3, Sb/TM6B, Tb | Bloomington Drosophila Stock Center | RRID: BDSC_56185 | y[1] w[*]; P{y[+t7.7] w[+mC]=UAS-mCD8.GFP.UAS-rCD2i}attP40 P{ry[+t7.2]=neoFRT}40 A/CyO; TM3, Sb[1]/TM6B, Tb[1] |
| Strain, strain background (D. melanogaster) | yw, hsFlp122; frt40a, UAS-CD2-RFP, UAS-GFP-miRNA | Gift from Claude Desplan Lab | yw, hsFLP; uas-CD2::RFP, UAS-GFP-miRNA/cyo; tm2/tm6b,tb | |
| Strain, strain background (D. melanogaster) | UAS-Notch-RNAi | Bloomington Drosophila Stock Center | RRID: BDSC_33611 | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS00001}attP2 |
| Strain, strain background (D. melanogaster) | UAS-Notch-ICD | Struhl and Greenwald, 2001 | ;UAS-Notch-ICD; +/+ | |
| Strain, strain background (D. melanogaster) | Hey-ORF-T2A-FlpD5 | Mark et al., 2021 | w; Hey-ORF-T2A-FLP(RFP+)/cyo, wg-LacZ; +/+ | |
| Strain, strain background (D. melanogaster) | UAS-FRT-STOP-FRT-myrGFP | Bloomington Drosophila Stock Center | RRID: BDSC_55810 | w[1118]; P{y[+t7.7] w[+mC]=10XUAS(FRT.stop)GFP.Myr}su(Hw)attP5 |
| Strain, strain background (D. melanogaster) | w; Brp-FRT-STOP-FRT-smGdP-v5-T2A-LexA; LexAop-Tomato | Peng et al., 2018; Xu et al., 2019 | w; Brp-FRT-STOP-FRT-smGdP-v5-T2A-LexA/cyo; LexAop-myrtdTomato/tm6b | |
| Strain, strain background (D. melanogaster) | UAS-Svp-RNAi | Bloomington Drosophila Stock Center | RRID: BDSC_28689 | y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF03105}attP2 |
| Strain, strain background (D. melanogaster) | 6–60 Gal4 | Gift from Lawrence Zipursky | W; Bl/cyo; 6–60 Gal4/tm6b | |
| Strain, strain background (D. melanogaster) | Bsh-L-Gal4 | Gift from Makoto Sato | ;Bsh-L-Gal4/cyo; +/+ | |
| Strain, strain background (D. melanogaster) | tubP-GAL80[ts] | Bloomington Drosophila Stock Center | RRID: BDSC_7017 | w[*]; P{w[+mC]=tubP-GAL80[ts]}2/TM2 |
| Strain, strain background (D. melanogaster) | UAS-Zfh1-RNAi | Vienna Drosophila Resource Center | VDRC 103205 | P{KK109931}VIE-260B |
| Strain, strain background (D. melanogaster) | 31C06-Gal4, UAS-myristoylated-tdTomato | Gift from Lawrence Zipursky | ;Bl/cyo; 31c06-Gal4, UAS- myristoylated-tdTomato/tm6b | |
| Strain, strain background (D. melanogaster) | E(spl)-mγ-GFP | Gift from Sarah Bray (Almeida and Bray, 2005) | w; +/+; E(spl)mγ-GFP/tm6b | |
| Strain, strain background (D. melanogaster) | VALIUM20-mCherry | Bloomington Drosophila Stock Center | RRID: BDSC_35785 | y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=VALIUM20-mCherry}attP2 |
| Strain, strain background (D. melanogaster) | Bsh-ORF-3XHA (86Fb) | FlyORF Webshop | Cat#F000054 | M{UAS-bsh.ORF.3xHA.GW}ZH-86Fb |
| Strain, strain background (D. melanogaster) | flyORF-TaDa | Bloomington Drosophila Stock Center | RRID: BDSC_91637 | w[1118]; M{RFP[3xP3.PB] w[+mC]=FlyORF-TaDa}ZH-86Fb |
| Strain, strain background (D. melanogaster) | hs-FlpD5; FlyORF-TaDa | Bloomington Drosophila Stock Center | RRID: BDSC_91638 | w[1118]; P{y[+t7.7] w[+mC]=hs-FLPD5}attP40; M{RFP[3xP3.PB] w[+mC]=FlyORF-TaDa}ZH-86Fb |
| Strain, strain background (D. melanogaster) | Bsh-TaDa | Xu et al., 2023 | w; +/CyO; UAS-GFP-Bsh-DAM/tm6b | |
| Antibody | Chicken polyclonal | Abcam | Cat#ab13970, RRID_300798 | Anti-GFP (1:1000) |
| Antibody | Rabbit polyclonal | Medical & Biological Laboratories Co. | Code#PM005 | Anti-RFP (1:500) |
| Antibody | Rabbit polyclonal | Gift from Claude Desplan (Özel et al., 2021) | Anti-Bsh (1:1000) | |
| Antibody | Guinea pig polyclonal | Gift from Lawrence Zipursky (Tan et al., 2015) & Makoto Soto | Anti-Bsh (1:1000) | |
| Antibody | Rat monoclonal | This study | Anti-Bsh (1:750); see methods section- generating Bsh antibody | |
| Antibody | Rabbit polyclonal | Gift from Markus Affolter (Bieli et al., 2015) | Anti-Apterous (1:1000) | |
| Antibody | Rat monoclonal | Gift from Cheng-Ting Chien (Chen et al., 2012) | Anti-Pdm3 (1:200) | |
| Antibody | Rabbit polyclonal | Gift from Cheng-Yu Lee (Janssens et al., 2014) | Anti-Erm (1:100) | |
| Antibody | Rat monoclonal | Gift from Jing Peng (Santiago et al., 2021) | Anti-Erm (1:70) | |
| Antibody | Mouse monoclonal | Developmental Studies Hybridoma Bank | Cat#Seven-up D2D3, RRID_2618079 | Anti-Svp (1:10) |
| Antibody | Rabbit polyclonal | Gift from James Skeath (Tian et al., 2004) | Anti-Zfh1 (1:1000) | |
| Antibody | Rabbit polyclonal | Asian Distribution Center for Segmentation Antibodies | Code#812 | Anti-Tailless (1:200) |
| Antibody | Mouse monoclonal | Developmental Studies Hybridoma Bank | Cat#Elav-9F8A9, RRID: AB_528217 | Anti-Elav (1:200) |
| Antibody | Mouse monoclonal | Bio-Rad Laboratories | Cat#MCA1360A647, RRID: AB_770156 | Anti-V5-TAG:Alexa Fluor 647 (1:300) |
| Antibody | Mouse monoclonal | Developmental Studies Hybridoma Bank | Cat#nc-82, RRID: AB_2314866 | Anti-Brp (1:50) |
| Antibody | Mouse monoclonal | Developmental Studies Hybridoma Bank | Cat#C594.9B, RRID: AB_528194 | Anti-Delta (1:10) |
| Antibody | Mouse monoclonal | TaKaRa | Cat#632543 | Anti-Cherry (1:500) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat# 711-475-152 AB_2340616 | DyLight 405 anti-rabbit (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#703-545-155, RRID: AB_2340375 | Alexa Fluor 488 anti-chicken (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#706-545-148, RRID: AB_2340472 | Alexa Fluor 488 anti-guinea pig (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#711-545-152, RRID: AB_2313584 | Alexa Fluor 488 anti-rabbit (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#715-545-150, RRID: AB_2340846 | Alexa Fluor 488 anti-mouse (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#715-295-151, RRID: AB_2340832 | Rhodamine Red-X anti-mouse (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#712-295-153, RRID: AB_2340676 | Rhodamine Red-X anti-rat (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#711-295-152, RRID: AB_2340613 | Rhodamine Red-X anti-rabbit (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#706-295-148, RRID: AB_2340468 | Rhodamine Red-X donkey anti-guinea pig (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#711-605-152, RRID: AB_2492288 | Alexa Fluor 647 donkey anti-rabbit (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#715-605-151, RRID: AB_2340863 | Alexa Fluor 647 donkey anti-mouse (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#706-605-148, RRID: AB_2340476 | Alexa Fluor 647 anti-guinea pig (1:400) |
| Antibody | Donkey polyclonal | Jackson ImmunoResearch Lab | Cat#712-605-153, RRID: AB_2340694 | Alexa Fluor 647 anti-rat (1:400) |
| Sequence-based reagent | Oligonucloetide | Integrated DNA technologies | DamID Adaptor (top strand): CTAATACGACTCACTATAGGGCAGCGTGGTCGCGGCCGAGGA | |
| Sequence-based reagent | Oligonucloetide | Integrated DNA technologies | DamID Adaptor (bottom strand): TCCTCGGCCG | |
| Sequence-based reagent | Oligonucloetide | Integrated DNA technologies | DamID Primer for PCR: GGTCGCGGCCGAGGATC | |
| Commercial assay or kit | QIAamp DNA Micro Kit | Qiagen | Cat#56304 | |
| Commercial assay or kit | PCR Purification Kit | Qiagen | Cat#28104 | |
| Chemical compound, drug | EDTA | Sigma-Aldrich | Cat#E6758 | |
| Chemical compound, drug | DpnI and CutSmart buffer | NEB | Cat#R0176S | |
| Chemical compound, drug | DpnII and DpnII buffer | NEB | Cat#R0543S | |
| Chemical compound, drug | MyTaq HS DNA Polymerase | Bioline | Cat#BIO-21112 | |
| Chemical compound, drug | AlwI | NEB | Cat#R0513S | |
| Chemical compound, drug | RNase A (DNase free) | Roche | Cat#11119915001 | |
| Chemical compound, drug | T4 DNA ligase and 10 x buffer | NEB | Cat#M0202S | |
| Software, algorithm | Fiji | Schindelin et al., 2012 | https://imagej.nih.gov/ij/download.html | |
| Software, algorithm | FastQC (v0.11.9) | The Babraham Bioinformatics group | https://www.bioinformatics.babraham.ac.uk/projects/download.html#fastqc | |
| Software, algorithm | damidseq_pipeline | Marshall and Brand, 2015 | https://owenjm.github.io/damidseq_pipeline/ | |
| Software, algorithm | Bowtie2 (v2.4.5) | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | |
| Software, algorithm | IGV (v.2.13.2) | Robinson et al., 2011 | https://software.broadinstitute.org/software/igv/download | |
| Software, algorithm | SAMtools (v1.15.1) | Li et al., 2009 | http://www.htslib.org/download/ | |
| Software, algorithm | deepTools (v3.5.1) | Ramírez et al., 2016 | https://deeptools.readthedocs.io/en/develop/content/installation.html | |
| Software, algorithm | Find_peaks | Marshall et al., 2016 | https://github.com/owenjm/find_peaks; (Marshall, 2016) |
Additional files
-
Supplementary file 1
Comparison of open chromatin regions between L4 and L5 neurons.
- https://cdn.elifesciences.org/articles/90136/elife-90136-supp1-v1.xlsx
-
Supplementary file 2
Genes that are expressed in L4 neurons but not L5.
- https://cdn.elifesciences.org/articles/90136/elife-90136-supp2-v1.xlsx
-
Supplementary file 3
Genes that are expressed in L5 neurons but not L4.
- https://cdn.elifesciences.org/articles/90136/elife-90136-supp3-v1.xlsx
-
Supplementary file 4
Comparison of Bsh genome-binding loci between L4 and L5 neurons.
- https://cdn.elifesciences.org/articles/90136/elife-90136-supp4-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/90136/elife-90136-mdarchecklist1-v1.pdf