The PCNA unloader Elg1 promotes recombination at collapsed replication forks in fission yeast
Figures
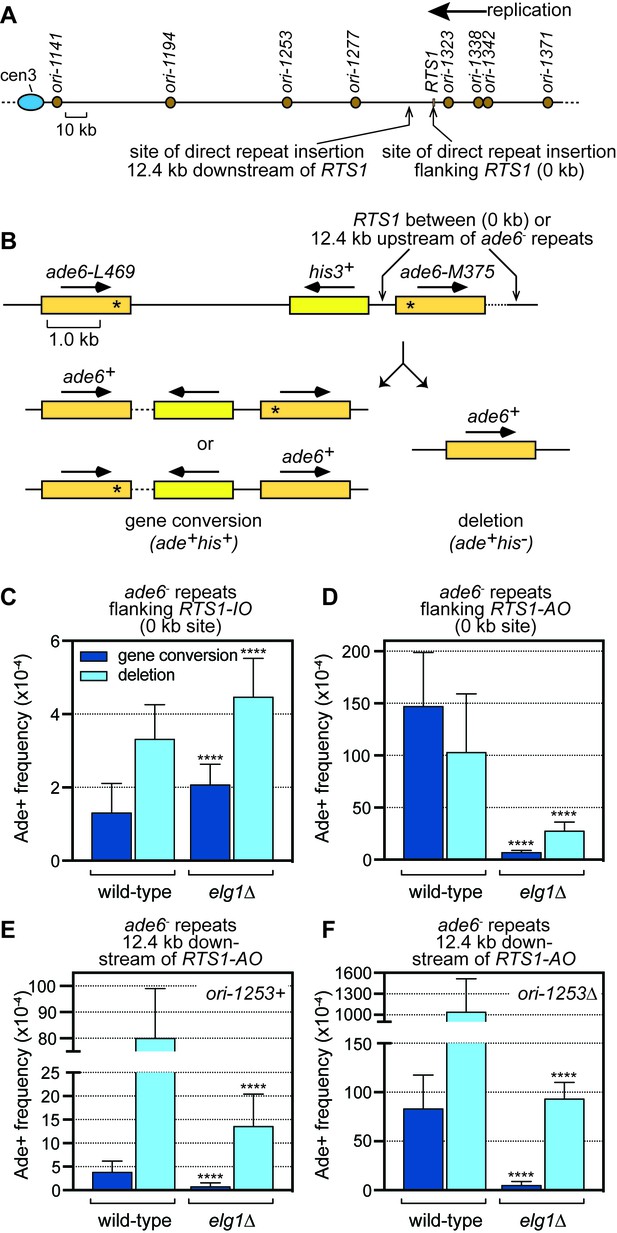
Spontaneous (RTS1-IO) and RTS1-AO-induced direct repeat recombination in wild-type and elg1∆ strains.
(A) Map showing the position of RTS1, origins of replication and sites of insertion for the direct repeat recombination reporter on chromosome 3. (B) Schematic of the direct repeat recombination reporter showing the ‘0 kb’ and ‘12.4 kb’ RTS1 sites and two types of Ade+ recombinant. Asterisks indicate the position of point mutations in ade6-L469 and ade6-M375. (C) Ade+ recombinant frequencies for strains MCW4712 and MCW7706. (D) Ade+ recombinant frequencies for strains MCW4713 and MCW7708. (E) Ade+ recombinant frequencies for strains MCW7259 and MCW8191. (F) Ade+ recombinant frequencies for strains MCW7295 and MCW8290. Data are mean values with error bars showing 1 SD. Significant differences relative to equivalent wild-type values are indicated by *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Ade+ recombinant frequencies with statistical analysis are also shown in Supplementary file 1.
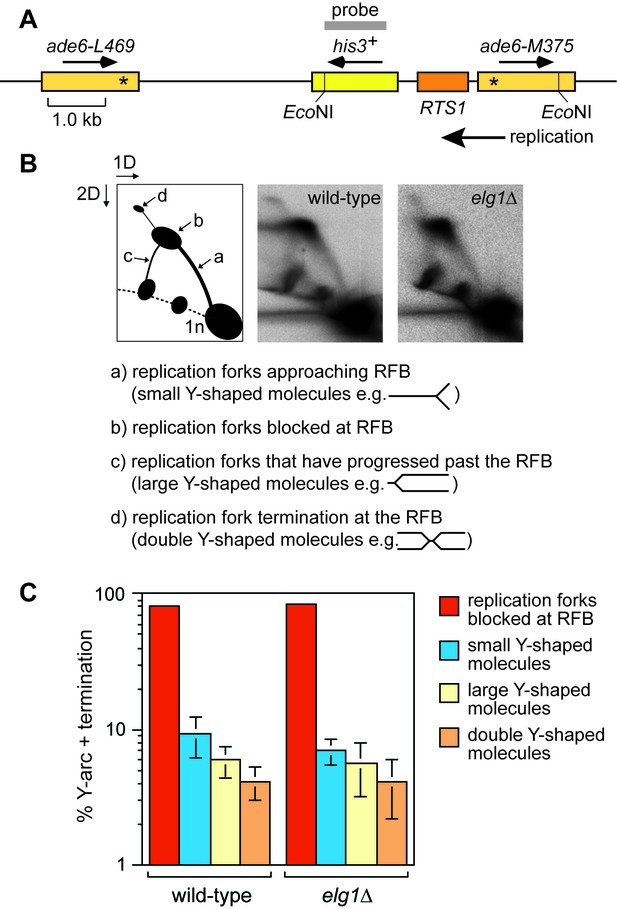
Elg1 is not required for replication fork blocking at RTS1-AO.
(A) Schematic showing the EcoNI restriction fragment and probe used for the 2DGE analysis in B. (B) 2DGE of replication intermediates in the EcoNI fragment shown in A. The DNA was extracted from strains MCW4713 and MCW7708. (C) Quantification of 2DGE. Data are mean values ± SD from three independent experiments.
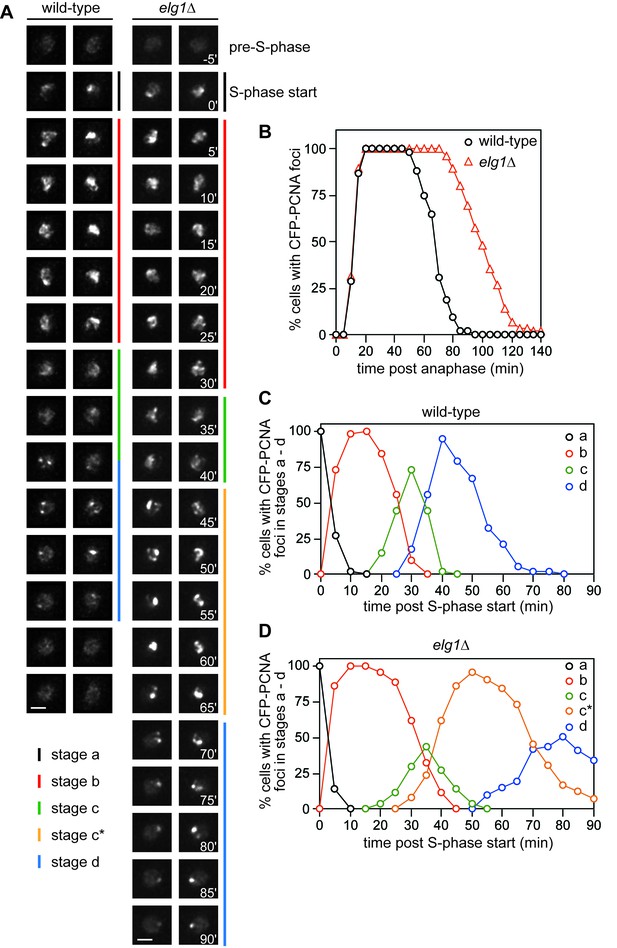
Comparison of CFP-PCNA fluorescence in wild-type and elg1∆ strains.
(A) Stills taken from four separate time-lapse movies (two each for wild-type and elg1∆) showing the different stages of nuclear CFP-PCNA fluorescence at 5 min intervals during representative cell cycles. Scale bar = 2 µm. (B) The percentage of cells with nuclear CFP-PCNA fluorescence above the pre-S-phase level at the indicated times post anaphase. (C – D) The percentage of cells with nuclear CFP-PCNA fluorescence in stages a – d. The data in B – D are taken from time-lapse movies like those shown in A. The wild-type strain is MCW7065 (n = 52) and the elg1∆ strain is MCW7965 (n = 85).
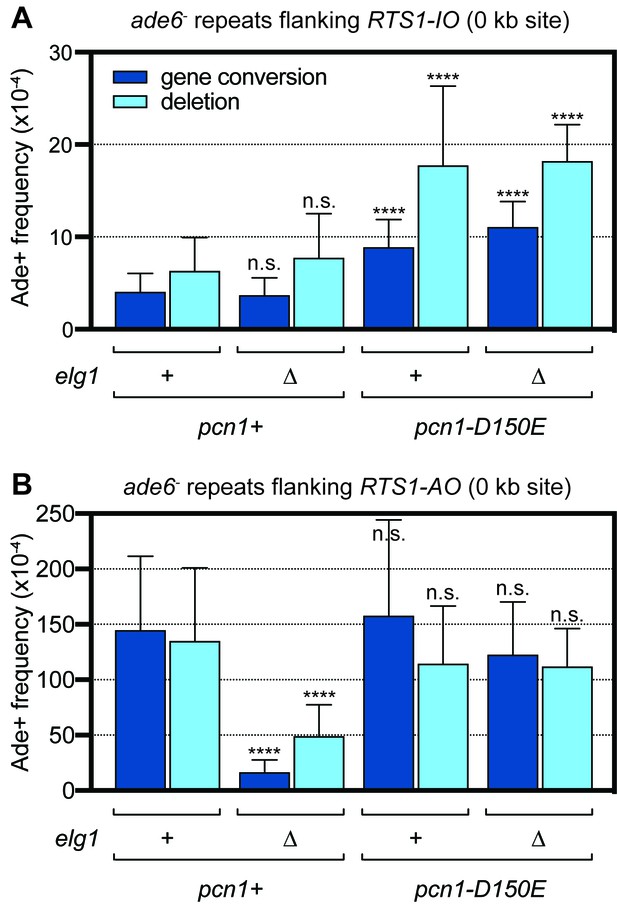
Spontaneous (RTS1-IO) and RTS1-AO-induced direct repeat recombination in pcn1+ and pcn1D150E strains with and without elg1.
(A) Ade+ recombinant frequencies for strains MCW9394, MCW9390, MCW9183 and MCW9187. (B) Ade+ recombinant frequencies for strains MCW9396, MCW9392, MCW9185 and MCW9189. Data are mean values with error bars showing 1 SD. Significant differences relative to equivalent elg1+ pcn1+ values are indicated by *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. n.s. = not significant. Ade+ recombinant frequencies with statistical analysis are also shown in Supplementary file 1.
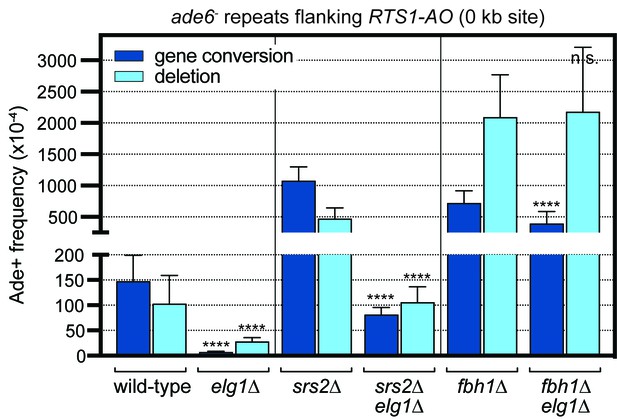
Effect of deleting srs2 and fbh1 on the frequency of RTS1-AO-induced direct repeat recombination in wild-type and elg1∆ strains.
The strains are MCW4713, MCW7708, FO1750, MCW8330, FO1816 and MCW8946. Data are mean values with error bars showing 1 SD. Significant differences relative to equivalent elg1+ strain values are indicated by *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. n.s. = not significant. Ade+ recombinant frequencies with statistical analysis are also shown in Supplementary file 1.
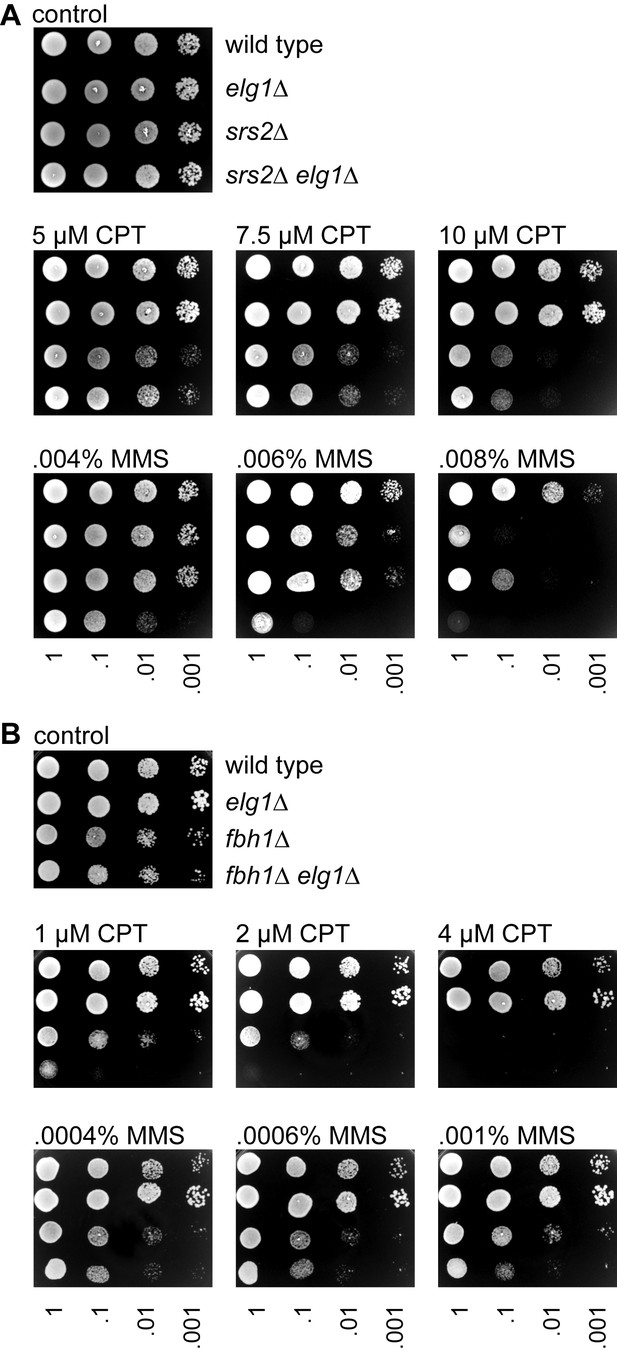
Growth and genotoxin sensitivities of elg1∆, srs2∆ and fbh1∆ single and double mutants.
(A) Spot assay comparing the growth, and CPT and MMS sensitivities of wild-type (MCW1221), elg1∆ (MCW7586), srs2∆ (MCW1017) and srs2∆ elg1∆ (MCW8332) strains. (B) Spot assay comparing the growth, and CPT and MMS sensitivities of wild-type (MCW4713), elg1∆ (MCW7708), fbh1∆ (FO1816) and fbh1∆ elg1∆ (MCW8946) strains. Assays were repeated at least once to ensure reproducibility.
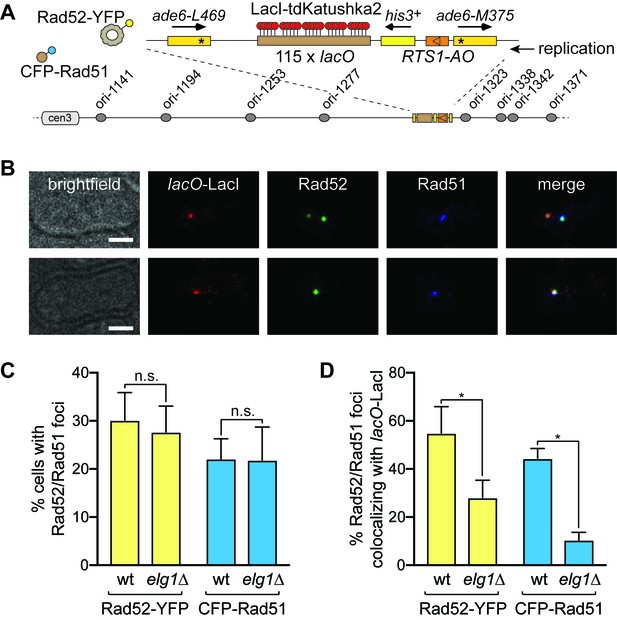
CFP-Rad51 and Rad52-YFP foci colocalization with RTS1-AO in wild-type and elg1∆ strains.
(A) Schematic showing key components of the strains used for imaging, including the modification of the 0 kb site recombination reporter with a 115 repeat lacO array. (B) Examples of two wild-type cells with CFP-Rad51 and Rad52-YFP foci. In the top panels, the cell contains two Rad52-YFP foci, one of which co-localizes with a lacO-LacI-tdKatushka2 focus, which marks the RTS1-AO location, and the second co-localizes with a CFP-Rad51 focus. The bottom panels show a cell with CFP-Rad51 and Rad52-YFP foci co-localizing with a lacO-LacI-tdKatushka2 focus. The scale bar = 2 µm. (C) Percentage of wild-type and elg1∆ cells with Rad52-YFP and CFP-Rad51 foci. Note that the values for wild-type are higher than observed previously (Nguyen et al., 2015) due to a greater proportion of S-phase and early G2 cells in the cultures that were analysed (data not shown). (D) Percentage of Rad52-YFP/CFP-Rad51 foci positive cells containing a Rad52-YFP and/or CFP-Rad51 focus that co-localizes with lacO-LacI-tdKatushka2. Data in (C) and (D) are mean values from four independent experiments (~100 cells were analysed in each experiment). Error bars show 1 SD. Significant differences between wild-type and elg1∆ are indicated by *p<0.05. n.s. = not significant. The strains are MCW7638 (wild-type) and MCW8921 (elg1∆).
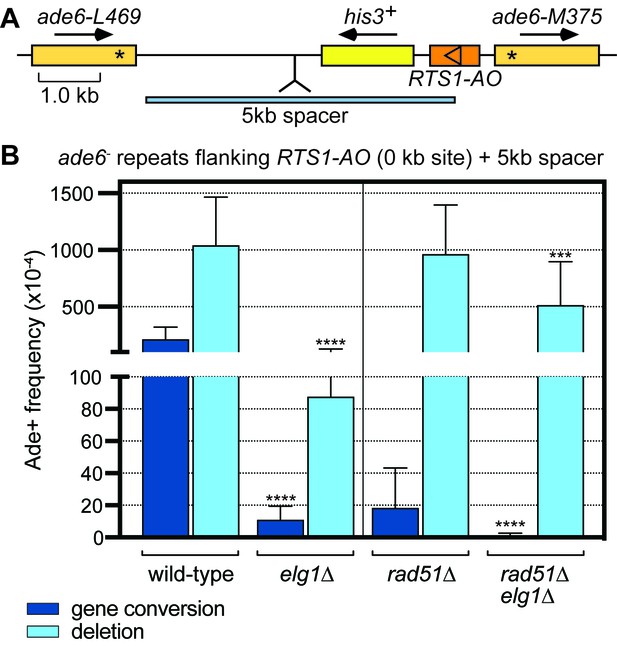
Elg1 promotes IFSA in the presence and absence of Rad51.
(A) Schematic showing the insertion of the 5 kb DNA spacer at the 0 kb site recombination reporter. (B) Ade+ recombinant frequencies for strains MCW8023, MCW8941, MCW8136 and MCW8943. Data are mean values with error bars showing 1 SD. Significant differences relative to equivalent elg1+ strain values are indicated by *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Ade+ recombinant frequencies with statistical analysis are also shown in Supplementary file 1.
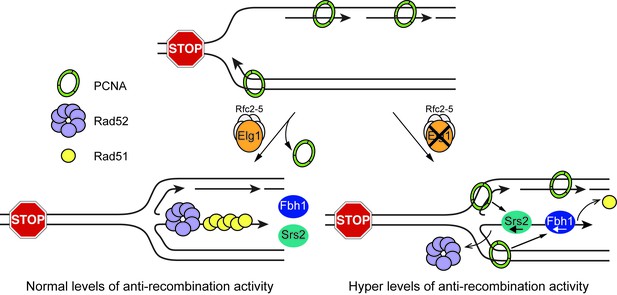
Model showing how Elg1 promotes recombination at a blocked replication fork.
Upon encounter with RTS1-AO, the replication fork undergoes reversal forming a ‘chicken foot’ structure with an exposed duplex DNA end. Resection of this DNA duplex, by the nucleolytic activity of the Mre11-Rad50-Nbs1-Ctp1 complex and Exo1, generates a 3’-ended ssDNA tail onto which Rad51 loads with the help of Rad52. The loading and/or retention of Rad51 and Rad52 at the barrier is inhibited by the activities of Fbh1 and/or Srs2, which are promoted either directly or indirectly by PCNA bound to the blocked fork. Elg1 unloads PCNA from the blocked fork and thereby limits Fbh1 and Srs2 activity. In the absence of Elg1, PCNA is retained at the blocked fork leading to excessive Fbh1 and Srs2 activity and, therefore, reduced recombination.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (S. pombe) | various strains | Ahn et al., 2005 | standard laboratory strain (972) derivatives; see Supplementary file 2 | |
| Strain, strain background (S. pombe) | various strains | Nguyen et al., 2015 | standard laboratory strain (972) derivatives; see Supplementary file 2 | |
| Strain, strain background (S. pombe) | various strains | this paper | standard laboratory strain (972) derivatives; see Supplementary file 2 | |
| Strain, strain background (S. pombe) | various strains | Lorenz et al., 2009 | standard laboratory strain (972) derivatives; see Supplementary file 2 | |
| Strain, strain background (S. pombe) | various strains | Morrow et al., 2017 | standard laboratory strain (972) derivatives; see Supplementary file 2 | |
| Recombinant DNA reagent | pMW777 | this paper | plasmid; see Materials and methods | |
| Recombinant DNA reagent | pST2 | this paper | plasmid; see Materials and methods | |
| Sequence-based reagent | oMW1884 | this paper | oligonucleotide; see Materials and methods | |
| Sequence-based reagent | oMW1885 | this paper | oligonucleotide; see Materials and methods |
Additional files
-
Supplementary file 1
Direct repeat recombinant frequencies.
- https://doi.org/10.7554/eLife.47277.011
-
Supplementary file 2
Schizosaccharomyces pombe strains.
- https://doi.org/10.7554/eLife.47277.012
-
Transparent reporting form
- https://doi.org/10.7554/eLife.47277.013





