De novo design of a homo-trimeric amantadine-binding protein
Figures
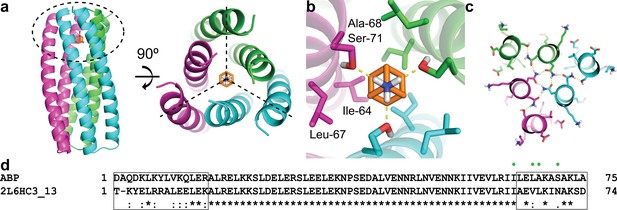
Computational design methodology.
(a) The homo-trimeric scaffold was designed to bind amantadine such that the C3 axes of the protein and the small molecule are aligned. Amantadine is colored orange and each monomer of ABP is colored magenta, green, or cyan. The amantadine binding site is highlighted by a dashed oval. (b) The binding pocket in ABP was designed to have polar serine residues (Ser-71) that hydrogen-bond (yellow dashed lines) to the amino group of amantadine and nonpolar residues (Ile-64, Leu-67, and Ala-68) to complement the shape of the hydrophobic moiety of amantadine. (c) The design model contains hydrogen-bond networks that specify the trimeric assembly of ABP. (d) A sequence alignment of ABP and 2L6HC3_13 is shown, with mutated regions shown in black boxes. Both sequences are preceded by a five-residue GHSMG pre-sequence (not shown) that result from the cloning strategy. The residues highlighted in (b) are annotated by green circles.

ABP variant designs.
(a) The original ABP design is shown colored in magenta, green, or cyan. (b) Ala/Ser mutations were introduced in the core of the helical bundle in order to reduce the trimeric interface. (c) HBNet was used to incorporate more polar residues at the monomer-monomer interfaces that could make hydrogen-bond interactions (yellow dashed lines). (d-f) The outer helices were truncated to various lengths and the identities of surface-exposed positions redesigned to be polar. (g) The helical bundle was shortened by removing a heptad repeat. (h) The helical bundle was shortened to just the amantadine-binding site, and helical repeats were fused onto the core bundle in order to stabilize the structure of the reduced core.
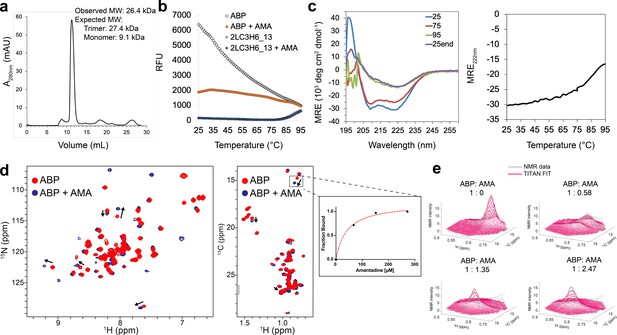
Binding characterization of amantadine to ABP.
(a) SEC chromatogram monitoring absorbance at 280 nm (mAU) and estimated molecular mass (from MALS). (b) Apo-ABP (orange, open circle) exhibits a high initial fluorescence signal that is lowered in the presence of amantadine (orange, solid circle). As expected, 2LC3H6_13 (blue, open triangle) and 2LC3H6_13 plus amantadine (blue, solid triangle) exhibit a very low initial fluorescence signal and overlap almost identically. (c) The CD spectrum of ABP at 25°C, 75°C, 95°C, and 25°C after heating and cooling. The CD spectrum of ABP at 25°C suggests an all ɑ-helical structure that remains fairly stable up to 75°C. (d) 2D amide 1H-15N HMQC spectra (left) and 2D methyl 1H-13C HMQC spectra (right) of 250 μM apo-ABP (red) or ABP in the presence of 2 mM amantadine (blue) recorded at 800 MHz, 37°C. Titration of amantadine leads to significant changes in the ABP NMR spectra (arrows). To the right of the 2D methyl methyl 1H-13C HMQC spectra an inset of dissociation constant estimate through conventional fraction bound analysis is shown for the affected ILE methyl group, with an estimated KD of <55 μM. (e) NMR line shape fitting of ABP throughout the NMR titration with amantadine performed in the program TITAN using a two-state binding model for the affected ILE methyl group. The NMR data (gray) are shown versus the TITAN fit (magenta).

CD spectrum of ABP in the presence amantadine.
(a) The CD spectrum of ABP in the presence of 5 mM amantadine at 25°C, 75°C, 95°C, and 25°C after heating and cooling suggests that the thermal stability of ABP is not significantly affected by the presence of amantadine. (b) CD temperature melt monitoring absorption at 222 nm for ABP in the presence of 5 mM amantadine.
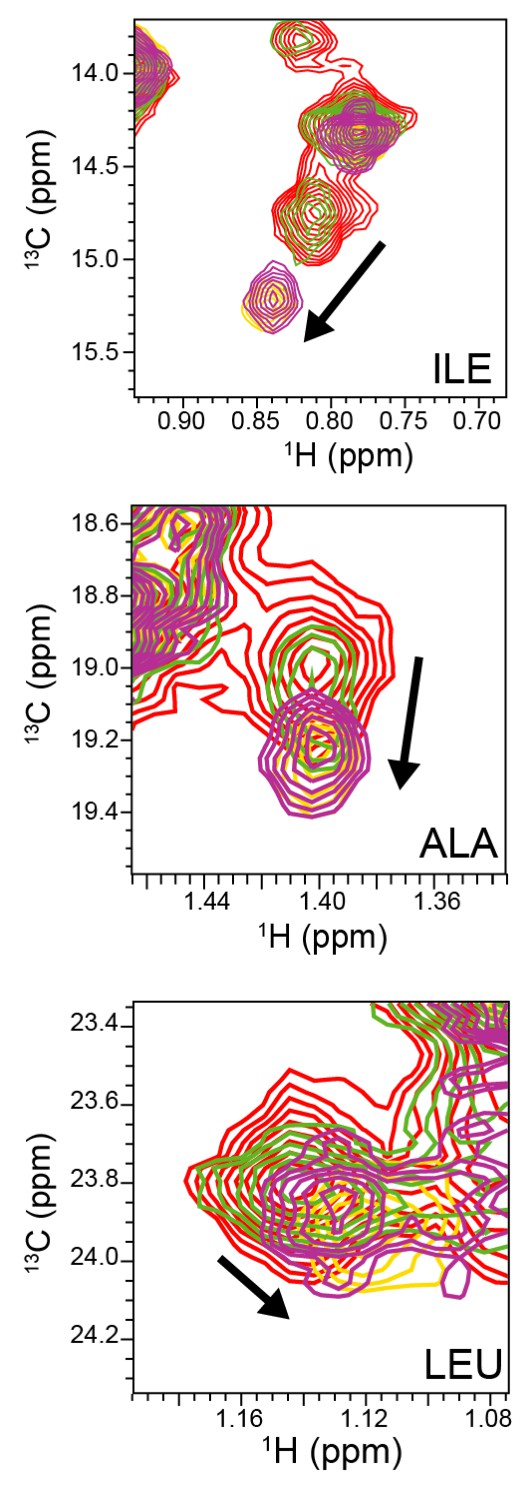
NMR titration of ABP with amantadine.
Representative view of 2D 1H-13C methyl SOFAST-HMQC spectra of select ABP methyl groups without amantadine (red) and with amantadine at ABP:amantadine molar ratios of 1:0.58 (green), 1:1.35 (purple), and 1:2.47 (yellow). Arrows indicate significant chemical shift deviations in ABP methyl groups upon complex formation with amantadine. NMR experiments were recorded with 118 μM ABP at 800 MHz, 37°C.
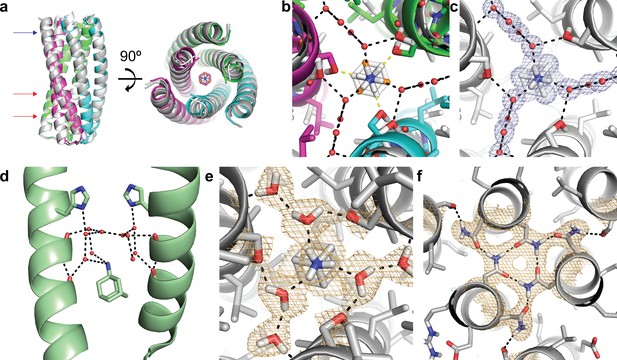
Structural characterization of the ABP-amantadine interaction.
(a) The high-resolution X-ray structure (white) and neutron structure (gray) of ABP in complex with amantadine are very close to the computational model (magenta, green, and cyan) (RMSD of 0.63 Å and 0.59 Å, respectively). The blue arrow indicates the amantadine binding site shown in (c,e), and the red arrows indicate the hydrogen bond networks, one of which is shown in (f). (b) A zoomed in overlay of the X-ray structure (white) and the design model (colored) reveal a shift in the helices within the amantadine-binding region, accompanied by a ~ 60° rotation of amantadine and the presence of water molecules that that mediate hydrogen bonding to Ser-71. Yellow dashed lines show direct hydrogen bonds to Ser-71 in the design model and black dashed lines show the hydrogen bonds observed in the X-ray structure. (c) Clear electron density can be observed for amantadine and ordered water molecules in the binding site of ABP (2Fo - Fc map contoured at 1.0σ). Water-mediated hydrogen bonds are observed between Ser-71 and the amino group of amantadine (black dashed lines). (d) The crystal structure (pale green) of amantadine bound to the influenza M2 protein through water-mediated hydrogen bonds (image generated from PDB: 6BKK [Thomaston et al., 2018]). (e) The nuclear scattering length density map shows the positions of deuterium atoms, including two ordered water molecules that mediate the hydrogen-bond network between Ser-71 and amantadine (2Fo - Fc contoured at 1.0σ). Hydrogen bonds are shown as black dashed lines. (f) Clear nuclear scattering length density can be observed for residues involved in the designed hydrogen-bond networks (black dashed lines) in ABP (2Fo - Fc map contoured at 1.0σ).
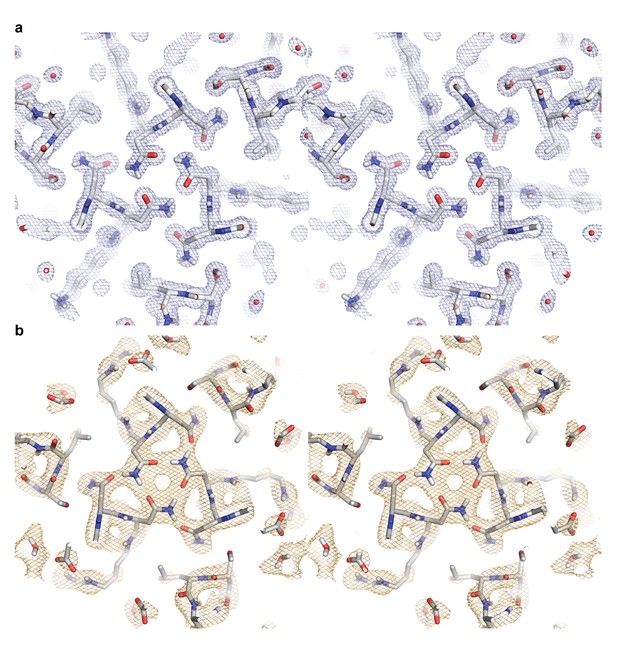
Stereo images of the electron and neutron length scattering density maps for a hydrogen bond network in ABP.
(a) The 2Fo - Fc electron density map contoured at 1.0σ is colored in light blue. (b) The 2Fo - Fc neutron density length scattering map contoured at 1.0σ is colored in light orange.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (include species and sex here) | One ShotBL21 Star (DE3) Chemically Competent E. coli | Invitrogen (Thermo Fisher Scientific) | C601003 | |
| Recombinant DNA reagent | pET28b(+) DNA - Novagen | Sigma-Aldrich (Millipore Sigma) | 69865–3 | |
| Commercial assay or kit | NeXtal Tubes JCSG+Suite | Qiagen | 130720 | |
| Chemical compound, drug | Amantadine hydrochloride | Sigma-Aldrich (Millipore Sigma) | A1260 | |
| Software, algorithm | Rosetta software suite | Rosetta Commons | N/A |
Additional files
-
Supplementary file 1
Supplementary file 1A RosettaScripts XML file (.xml).
Sample RosettaScripts XML file Supplementary file 1B Parameter constraint file for amantadine (.cst). Parameter constraint file for amantadine used in the RosettaDesign calculations. Supplementary file 1C Parameter definition file for amantadine (.params). Parameter definition file for amantadine used in the RosettaDesign calculations. Supplementary file 1D Restype file (in.res). Restype file used in the RosettaDesign calculations.
- https://cdn.elifesciences.org/articles/47839/elife-47839-supp1-v1.docx
-
Supplementary file 2
Supplementary file 2A X-ray data collection and refinement statistics.
Data collection and refinement statistics for the X-ray structure of ABP Supplementary file 2B Neutron scattering data collection and refinement statistics Data collection and refinement statistics for the neutron and room temperature X-ray structure of ABP.
- https://cdn.elifesciences.org/articles/47839/elife-47839-supp2-v1.docx
-
Supplementary file 3
NMR line shape fitting analysis with fixed KD values.
- https://cdn.elifesciences.org/articles/47839/elife-47839-supp3-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/47839/elife-47839-transrepform-v1.docx





