Plant Evolution: The inevitability of C4 photosynthesis
Understanding the evolution of complex innovations remains one of the most challenging problems in biology (Lynch, 2007; Wagner, 2014). Insights often stem from experimental lab studies that manipulate systems under 'directed evolution' (Weinreich et al., 2006; Blount et al., 2012; Finnigan et al., 2012). However, complex traits that have evolved many times over in independent lineages present a different—yet equally powerful—opportunity to infer the evolutionary trajectories of novel traits.
In flowering plants, C4 photosynthesis is a well-studied, complex adaptation that has independently evolved over 60 times (Sage et al., 2011). Many key, shared stages along the C4 evolutionary trajectory have been identified by studying multiple C4-evolving plant groups (e.g., Kennedy et al., 1980; Ku et al., 1983; Vogan et al., 2007; Williams et al., 2013). Now, in eLife, Udo Gowik and colleagues at Heinrich-Heine-Universität—including Julia Mallmann and David Heckmann as joint first authors—present a compelling new hypothesis for how the final evolutionary steps were realized (Mallmann et al., 2014).
Although atmospheric carbon dioxide (CO2) levels are currently rising, the last 30 million years witnessed great declines in CO2, which has limited the efficiency of photosynthesis. Rubisco, the critical photosynthetic enzyme that catalyses the fixation of CO2 into carbohydrate, also reacts with oxygen when CO2 levels are low and temperatures are high. When this occurs, plants activate a process known as photorespiration, an energetically expensive set of reactions that—importantly for this story—release one molecule of CO2.
C4 photosynthesis is a clever solution to the problem of low atmospheric CO2. It is an internal plant carbon-concentrating mechanism that largely eliminates photorespiration: a 'fuel-injection' system for the photosynthetic engine. C4 plants differ from plants with the more typical 'C3' photosynthesis because they restrict Rubisco activity to an inner compartment, typically the bundle sheath, with atmospheric CO2 being fixed into a 4-carbon acid in the outer mesophyll. This molecule then travels to the bundle sheath, where it is broken down again, bathing Rubisco in CO2 and limiting the costly process of photorespiration.
The evolution of the C4 pathway requires many changes. These include the recruitment of multiple enzymes into new biochemical functions, massive shifts in the spatial distribution of proteins and organelles, and a set of anatomical modifications to cell size and structure. It is complex, and it is also highly effective: C4 plants include many of our most important and productive crops (maize, sorghum, sugarcane, millet) and are responsible for around 25% of global terrestrial photosynthesis (Still et al., 2003).
A key intermediate step in the evolution of C4 is the establishment of a rudimentary carbon-concentrating mechanism. Termed 'C2 photosynthesis', this mechanism limits certain reactions of the photorespiratory cycle to the bundle sheath cells. A byproduct of these reactions is CO2, creating a slightly elevated CO2 concentration and increasing Rubisco efficiency in these cells. Though much rarer than C4 plants, C2 plants have been discovered in a variety of C4-evolving lineages, and are thought to represent a common, if not requisite, intermediate step along the C4 trajectory (Sage et al., 2012).
One implication of a restricted photorespiratory cycle is the development of a severe nitrogen imbalance between the mesophyll and the bundle sheath cells. This occurs because every molecule of CO2 produced in the bundle sheath is accompanied by a molecule of ammonia. While this nitrogen imbalance has previously been recognised (Monson and Rawsthorne, 2000), it has never been closely studied, and certainly never considered as potentially important to the evolutionary assembly of the C4 pathway.
To investigate this, Mallmann, Heckmann et al. combined a mechanistic model of C2 physiological function with a metabolic model, which allowed them to predict the buildup of certain metabolites based on the rates of Rubisco and photorespiratory activity. They then modelled the various biochemical pathways that could potentially be induced to balance metabolic fluxes between the mesophyll and bundle sheath cells. This creative combination of models allowed them to evaluate the various metabolic pathways for re-balancing nitrogen in terms of which pathways resulted in the highest biomass yield (a proxy for fitness).
Remarkably, when low levels of C4 enzyme activity are permitted in the model, key elements of the C4 cycle are favoured as the nitrogen-balancing pathway. What's more, this model predicts that with a C4 cycle established, increasing the activity of the enzymes results in a linear increase in biomass yield. Allowing for low levels of C4 enzyme activity is biologically reasonable, as these enzymes are routinely present in C3 leaves. Mallmann, Heckmann et al. support their model predictions with experimental gene expression data from a set of C3, C2, C4, and other C3-C4 intermediate types in the plant lineage Flaveria, which show elevated C4 cycle activity even in intermediates that are not using the enzymes to capture carbon.
In other words, once a C2 cycle is established, the evolution of a fully realized C4 process is fairly trivial. Once C4 enzymes are recruited to shuttle nitrogen back to the mesophyll, it is all but inevitable. This can explain in part why C4 has evolved such a startling number of times, and why many of these origins are highly clustered across the tree of life. Many C4 evolutionary clusters likely share an ancestor that had already acquired an elevated likelihood of evolving the pathway (Figure 1).
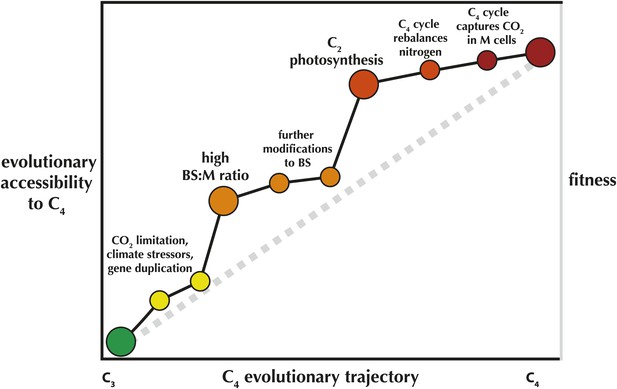
An 'Evolvability Landscape' for C4 photosynthesis.
Many intermediate stages along the evolutionary trajectory from C3 to C4 are well known (Sage et al., 2012). These can be displayed as part of an adaptive fitness landscape, which links biological properties (horizontal axis) with the fitness they produce (right vertical axis; a greater height indicates a greater fitness). The adaptive fitness landscape of the C4 trajectory was recently modelled as 'Mt. Fuji-like': a steep linear incline with each step along the trajectory bringing small, incremental increases in fitness (Heckmann et al., 2013), represented here by the grey dashed line. The gains in relative likelihood of evolving C4, or the 'evolutionary accessibility' of the pathway, may not be so linear (left vertical axis; black line). In spite of some limited flexibility in the order of trait acquisition (Williams et al., 2013), two intermediate stages are relatively fixed in position along the trajectory and also provide steep increases in C4 evolvability. One early step, an elevated ratio of bundle sheath: mesophyll cross-sectional area (BS:M ratio) was recently identified as a key trait that preceded multiple parallel realizations of C4 (Christin et al., 2013). Mallman et al. propose a mechanistic interaction between C2 and C4 photosynthesis, suggesting that evolution of the C2 stage of the trajectory greatly increases the probability that full C4 photosynthesis will quickly follow.
This may also explain why C2 species are so rare relative to C4 species—C2 is likely to be a step along the trajectory with a relatively short evolutionary lifespan. At the same time, it raises the question of why a handful of C2 species are persistent—the C2 Mollugo verticillata group may be up to 15 million years old (Christin et al., 2011). A testable hypothesis would be that these C2 plants have solved their nitrogen problem a different way, thereby limiting their own evolutionary accessibility to C4 photosynthesis. If so, this highlights the key role of contingency in adaptation, and our growing power to understand and predict macroevolutionary processes.
References
-
Anatomical enablers and the evolution of C4 photosynthesis in grassesProceedings of the National Academy of Sciences of the USA 110:1381–1386.https://doi.org/10.1073/pnas.1216777110
-
Complex evolutionary transitions and the significance of C3-C4 intermediate forms of photosynthesis in MolluginaceaeEvolution; International Journal of Organic Evolution 65:643–660.https://doi.org/10.1111/j.1558-5646.2010.01168.x
-
C3-C4 photosynthesis in the genus Mollugo: structure, physiology and evolution of intermediate characteristicsAmerican Journal of Botany 67:1207–1217.https://doi.org/10.2307/2442363
-
The frailty of adaptive hypotheses for the origins of organismal complexityProceedings of the National Academy of Sciences of the USA 104:8597–8604.https://doi.org/10.1073/pnas.0702207104
-
BookCO2 assimilation in C3-C4 intermediate plantsIn: Leegood R, Sharkey T, Caemmerer S, editors. Advances in photosynthesis and respiration. Dordrecht: Kluwer Academic. pp. 533–550.
-
The C4 plant lineages of planet EarthJournal of Experimental Botany 62:3155–3169.https://doi.org/10.1093/jxb/err048
-
Photorespiration and the evolution of C4 photosynthesisAnnual Review of Plant Biology 63:19–47.https://doi.org/10.1146/annurev-arplant-042811-105511
-
BookHomology, genes, and evolutionary innovationPrinceton, NJ: Princeton University Press.
Article and author information
Author details
Publication history
Copyright
© 2014, Edwards
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 3,404
- views
-
- 254
- downloads
-
- 5
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.





