The role of IMP dehydrogenase 2 in Inauhzin-induced ribosomal stress
Figures
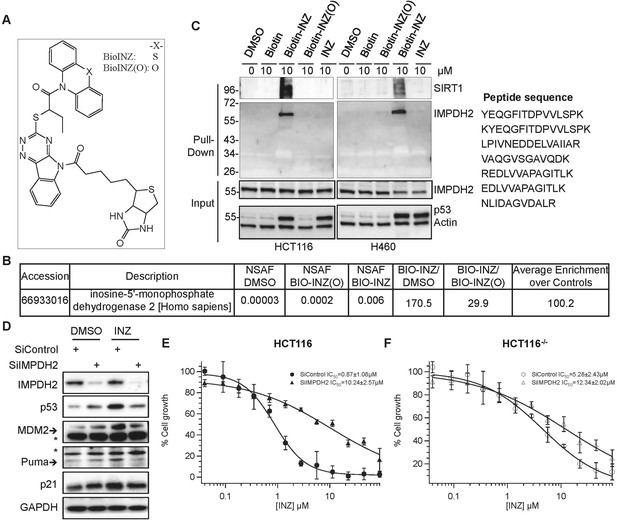
Identification of IMPDH2 as a potential target of INZ.
(A) Structure of INZ analogs conjugated with Biotin (Biotin-INZ) used for INZ target identification, the oxygen-substituent (Biotin-INZ (O)) as a negative control. (B–C) Cells were treated with indicated compounds individually for 18 hr. Cleared cell lysates were incubated with NeutrAvidin beads and washed. The samples from HCT116 cells were then in-beads digested for MS analysis. NSAF: normalized spectral abundance factor. Samples were also resolved by SDS-PAGE and subjected to IB with indicated antibodies. (D–F) Knockdown of IMPDH2 alleviates INZ induction of p53. HCT116 cells were transfected with scrambled siRNA or IMPDH2 siRNA. 18 hr prior to harvesting, cells were treated with 2 µM INZ and harvested for IB with indicated antibodies (D). HCT116 and HCT116−/− cells were exposed to INZ for 72 hr and evaluated by WST cell growth assays (E and F). The IC50 values of INZ in the scrambled siRNA and IMPDH2 siRNA transfected cells are 0.87 ± 1.08 µM and 10.24 ± 2.57 µM for HCT116 cells, and 5.28 ± 2.43 µM and 12.34 ± 2.02 µM for HCT116−/− cells, respectively (Mean ± SD, n = 3).
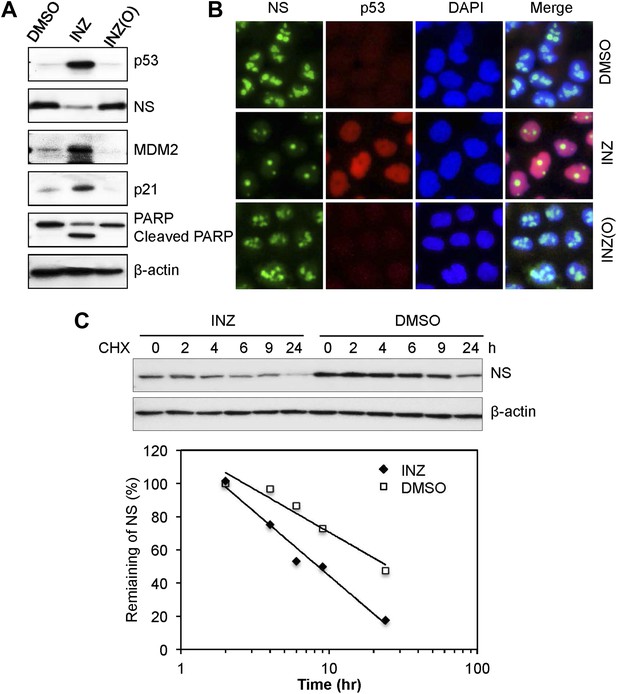
INZ, not its inactive analog INZ (O), treatment, decreases NS expression and destabilizes NS protein.
H460 cells were treated with 2 μM INZ or its analogue INZ (O) for 20 hr. Cells were harvested and immunoblotted with p53, NS, MDM2, p21, cleaved PARP and β-actin (A), or immunostained with anti-NS (green) and anti-p53 (red) (B). (C) H460 cells were treated with 2 μM INZ for 9 hr before then 50 μg/ml of CHX was added. Cells were harvested at different time points as indicated and assayed for levels of NS.
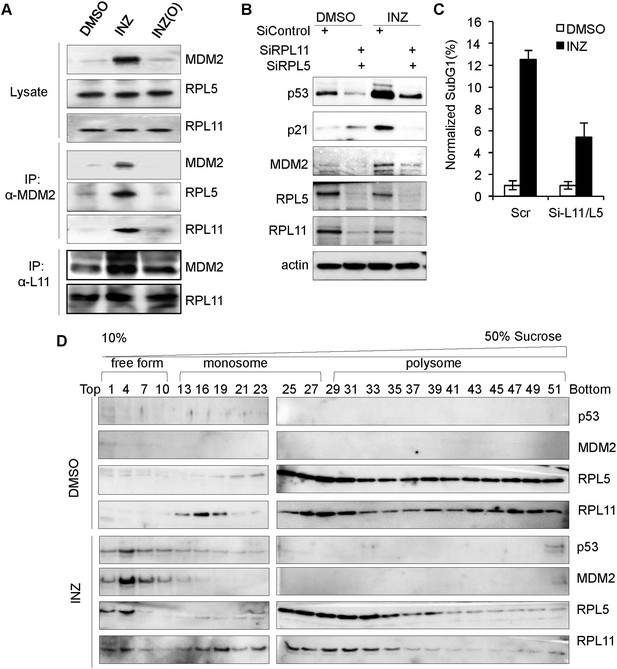
INZ treatment enhances the interaction of MDM2 with L5 and L11 by inducing ribosome-free form of RPL5 and RPL11.
(A) H460 cells were treated with 2 μM INZ or INZ (O) for 18 hr. Cell lysates were used for IP with anti-MDM2 antibodies or anti-L11 antibodies followed by IB using anti-RPL5, RPL11 or MDM2 antibodies. (B and C) HCT116 cells were transfected with siRNAs against RPL5 and RPL11, or control, for 48–72 hr and treated with 2 µM INZ 18 hr before harvesting, followed by IB using indicated antibodies or subG1 analysis by flow cytometry. (D) Ribosomal profile assay. Cytoplasmic extracts containing ribosomes from H460 cells treated with or without 2 µM INZ for 18 hr were subjected to a 10–50% linear sucrose gradient sedimentation centrifugation. Fractions were collected and subjected to IB with anti-RPL11, anti-RPL5, anti-p53, or anti-MDM2 antibodies. The distribution of ribosomes is indicated.
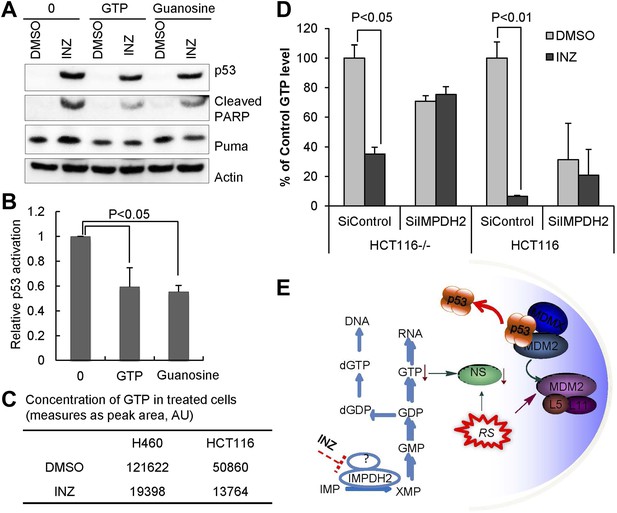
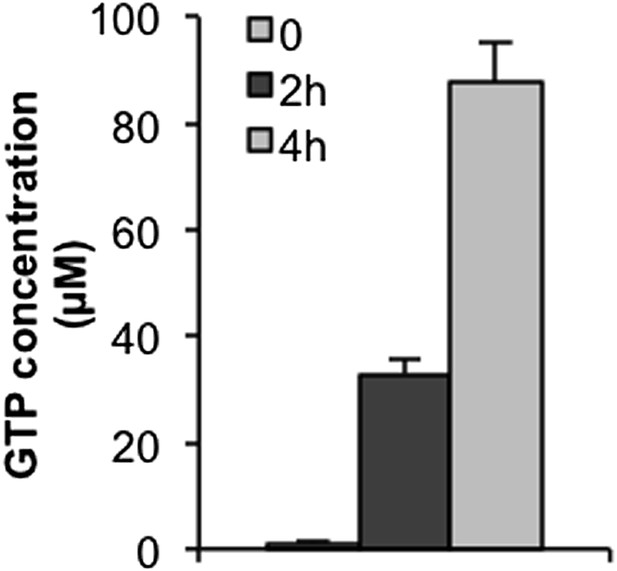
GTP or guanosine lessens INZ activation of p53 in cells.
(A and B) H460 cells were pretreated with GTP or guanosine for 2 hr before the addition of 2 μM INZ. Cells were harvested and followed by IB with indicated antibodies. (C) Effect of INZ on cellular GTP level. The nucleotides were extracted from H460 or HCT116 cells treated with 2 μM INZ by 80% acetonitrile and SPE column. Samples were subjected to GTP analysis by HPLC. Results of quantification of HPLC spectra presented in arbitrary units (AU) were presented in this Table. (D) HCT116−/− and HCT116 cells transfected IMPDH2 siRNA (SiIMPDH2) or scrambled siRNA (SiControl) were exposed to INZ for 18 hr, and cellular GTP was extracted, measured and quantitated by LC-MS/MS. Values represent means ±SD (n = 2). (E) A schematic diagram of the role of IMPDH2 in INZ-induced ribosomal stress (RS) and p53 activation. IMPDH2 is a rate-limiting enzyme in the de novo guanine nucleotide biosynthesis. INZ reduced the levels of cellular GTP and NS by targeting IMPDH2 (or its complex), resulting in RS that leads to the enhancement of the RPL11/RPL5-MDM2 interaction, consequently MDM2 inactivation and p53 activation.
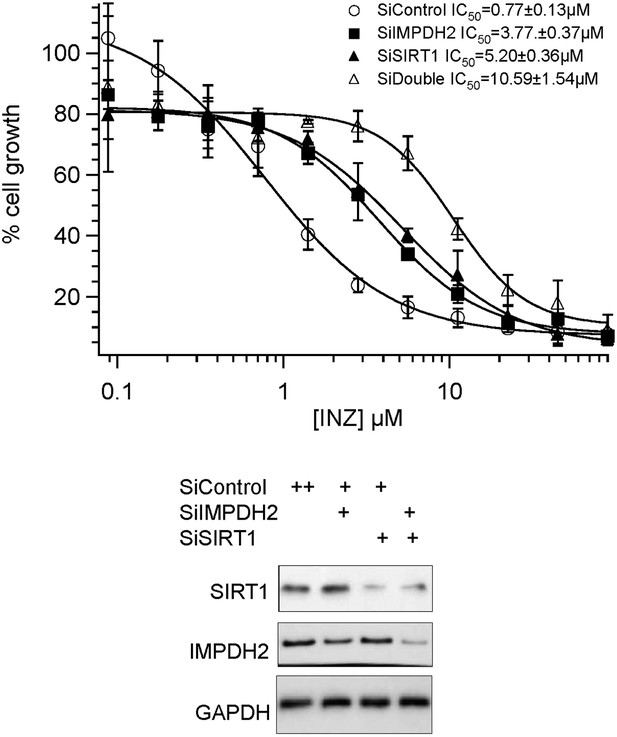
Combination effect of co-depletion of IMPDH2 and SIRT1 on INZ induced cell death. HCT116 cells, transfected with scrambled siRNA (SiControl), IMPDH2 siRNA (SiIMPDH2), SIRT1 siRNA (SiSIRT1) or co-transfected with IMPDH2 and SIRT1 siRNA, were treated with different doses of INZ and cell viability were assessed by WST cell growth assays. IC50 values are represented as mean ± standard deviation (n=3).
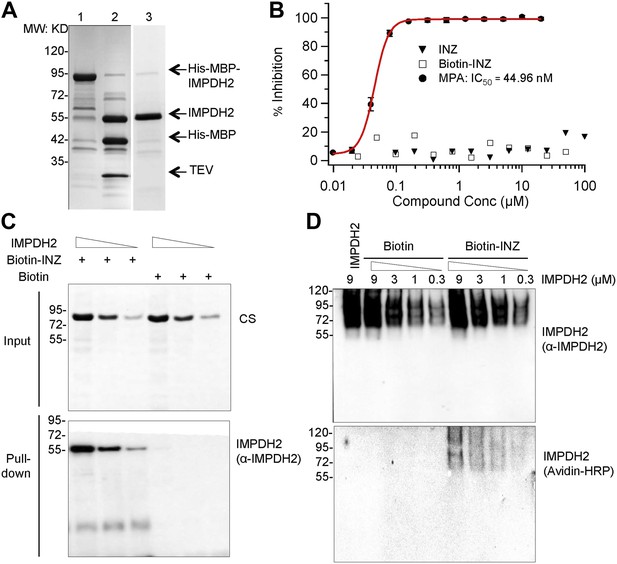
INZ binds to IMPDH2, but does not inhibit enzyme activity in vitro. (A) Recombinant IMPDH2 produced in E. coli BL21-CodonPlus (DE3)-RIPL, and purified through Ni-His columns (Lane 1) followed by TEV cleavage (Lane 2) as described in the experimental procedure. The purity of IMPDH2 enzyme (Lane 3) is confirmed before every assay by SDS-PAGE. (B) In vitro enzyme activity assays were conducted as described in the Supplementary Information. Mycophenolic Acid (MPA) is used as a positive control for IMPDH2 inhibition. The curve fitting and IC50 determination of INZ were performed using Igor Pro 4.01A. (C) Purified IMPDH2 was incubated at indicated concentrations with Biotin or Biotin-INZ that was conjugated with avidin beads. The bound IMPDH2 to Biotin-INZ was analyzed using IB with NeutrAvidin Protein-Horseradish Peroxidase Conjugated (Avidin-HRP, 1:1000; Pierce) and anti-IMPDH2 antibodies. (D) Purified IMPDH2 was incubated at indicated concentrations with Biotin-INZ or Biotin overnight at 4°C. After incubation, each mixture was subjected to Native-PAGE analysis, followed by blotting to PVDF. The blot was probed with Avidin-HRP and anti-IMPDH2 antibodies.